Spike S1 (B.1.1.7, Alpha Variant) (SARS-CoV-2): ACE2 Inhibitor Screening Colorimetric Assay Kit
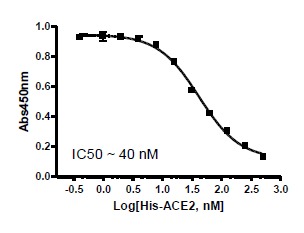
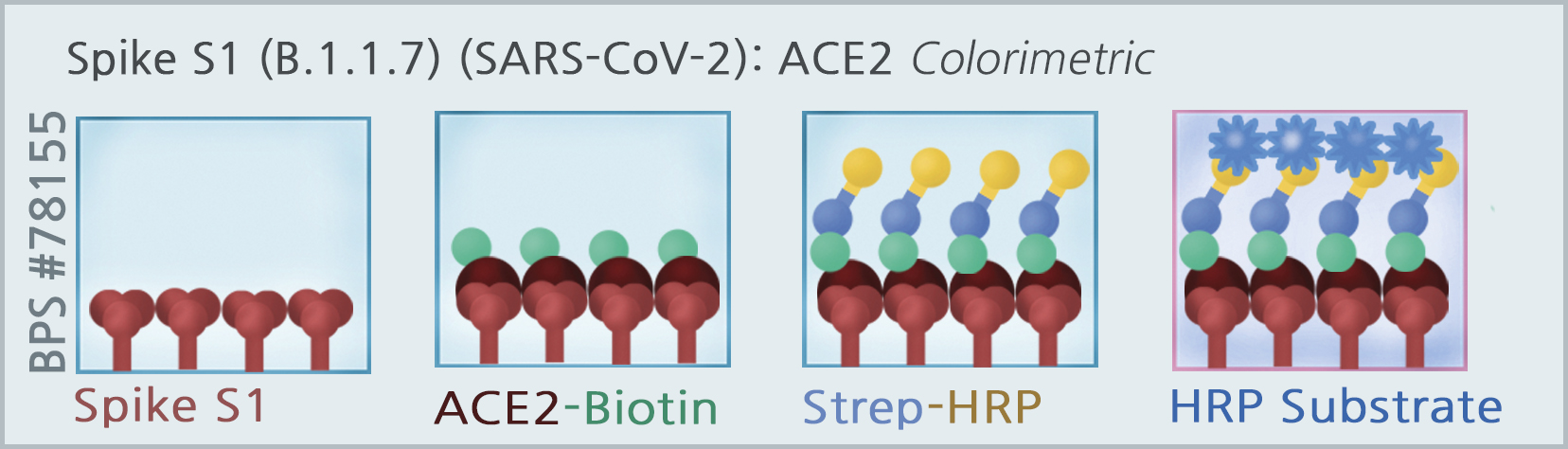
The Spike S1 (B.1.1.7 Variant) (SARS-CoV-2):ACE2 Inhibitor Screening Colorimetric Assay Kit is designed for screening and profiling inhibitors of the interaction of ACE2 with the B.1.1.7 variant of the SARS-CoV-2 Spike S1 protein. The key to this kit is the high sensitivity of detection of ACE2-Biotin protein by Streptavidin-HRP. Only a few simple steps on a microtiter plate are required for the assay. First, Spike S1 B.1.1.7 protein is coated on a 96-well transparent plate. Next, ACE2-Biotin is incubated with Spike S1 variant on the plate. Finally, the plate is treated with streptavidin-HRP followed by addition of an HRP substrate to produce color, which can then be measured using a UV/Vis spectrophotometer microplate reader.
Assay Principle
PBS (Phosphate buffered saline)
1N HCl (aqueous)
Rotating or rocker platform
UV/Vis spectrophotometer microplate reader capable of reading absorbance at 450 nm
| Catalog # | Name | Amount | Storage |
| Spike S1 (del_HV69-70, del_Y144, N501Y, A570D, D614G, P681H), His-tag (SARS-CoV-2) | 10 µg | -80°C | |
| 100665 | ACE2, His-Avi-Tag, Biotin-labeled HiP™ | 5 µg | -80°C |
| 79742 | Streptavidin-HRP | 10 µl | +4°C |
| 79311 | 3x Immuno Buffer 1 | 50 ml | -20°C |
| 79728 | Blocking Buffer 2 | 50 ml | +4°C |
| 79651 | Colorimetric HRP substrate | 10 ml | +4°C |
| 79964 | Transparent 96-well white microplate | 1 | Room Temp |
The pandemic coronavirus disease 2019 (COVID-19) is caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). As a first step of the viral replication strategy, the virus attaches to the host cell surface before entering the cell. The Spike protein of the SARS-CoV-2 recognizes and attaches to the Angiotensin-Converting Enzyme 2 (ACE2) receptor found on the surface of type I and II pneumocytes, endothelial cells, and ciliated bronchial epithelial cells. It has been widely suggested that active as well as passive immunizations targeting the interaction between the Spike protein of SARS-CoV-2 and ACE2 offer promising protection against the viral infection. However recent reports showed that a mutant strain first identified in the UK (B.1.1.7) exhibits higher transmissibility and infectivity.
The B.1.1.7 variant contains multiple mutations, including several in the Spike protein that leads to higher infectivity rates than the wild-type virus. The S1 subunit (a.a. 14-685) of the Spike protein includes the Receptor Binding Domain (RBD) region (a.a. 319-591) that is responsible for binding to the ACE2 receptor on target cells. Mutations outside of the RBD in the S1 subunit of spike are important for influencing the immunogenicity, conformation, and flexibility of the spike protein. Investigations on the effects of mutations on viral replication and pathogenesis will be critical for developing effective strategies for vaccines and antibody therapies against COVID-19.
1. Wang P. et al., Increased Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7 to Antibody Neutralization. bioRxiv 2021 Jan 26; 2021.01.25.428137
2. Shen X., et al., SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral Spike vaccines. bioRxiv. 2021 Jan 29; 2021.01.27.428516
3. Hoffman M. et al., SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020; 181:1-10