Targeted Protein Degradation Approaches and Applications
Introduction
Traditional approaches to regulate protein activity have relied on direct protein interactions by using inhibitors or ligands. However, these target only about 10% of the known 4000 disease-linked proteins, with many being considered “undruggable” due to their quaternary structure. In recent years new strategies to target “undruggable” proteins have been developed, harnessing the power of the cells’ own protein degradation machinery. These are collectively known as Targeted Protein Degradation (TPD) strategies. TPDs, contrary to inhibitors, work in a non-stoichiometric way, requiring the use of very small amounts of TPD, impact all the protein functions in the cell and do not tend to result in development of drug resistance (1). These characteristics represent major clinical advantages for cancer, neurodegenerative disease, inflammatory disease, and infection treatments. TPD has thus been an expanding field of research, particularly for pharmaceutical companies seeking to develop novel therapeutic strategies.
TPD Strategies and Clinical Applications
Eukaryotic cells have evolved highly complex and interlinked pathways to control the quality of proteins, from their synthesis to their degradation. An imbalance at any step of the proteostasis system may result in wild type or mutant protein accumulation, and finally disease. Defective proteins are targeted for removal by either the proteasome, via the ubiquitin-proteosome system (UPS), or by the lysosome. Targeting proteins for degradation involves ubiquitination of the protein, a multistep process involving E1 (Ub activating enzymes), E2 (Ub conjugating enzyme) and E3 Ligases, where one or several ubiquitins (Ub) are covalently attached to the target protein. The destination of Ub-protein depends on which lysine is ubiquitinated, with K48 poly-ubiquitination being targeted to the proteasome and K63 to the lysosome. The lysosome also digests organelles, protein aggregates and pathogens and receives the cellular “trash” via endocytosis, phagocytosis, or autophagy. Several strategies have been designed to take advantage of the proteasomal and lysosomal systems for disease-related protein removal, which include Proteolysis Targeting Chimeras (PROTAC®, a registered trademark of Arvinas, Inc.), molecular glues, and Lysosome-Targeting Chimera (LyTAC), amongst others (see Table).
PROTAC®
PROTACs were amongst the first TPD technologies to be developed that lead to protein degradation in the proteosome. They consist of a protein of interest (POI)-binding domain (peptide or small molecule), a linker and an E3 ligase ligand. The proximity of the POI to the E3 ligase in the ternary complex formed results in ubiquitin transfer to the POI and targeting for degradation. PROTACs have been successfully developed for a diverse range of proteins involved in cancer, including kinases, transcription factors and GTPases.
Examples of PROTACs targeting BTK (Bruton tyrosine kinase), p38 and BCL-xL, demonstrate additional advantages of this technology. BTK is involved in chronic lymphocytic leukemia, and a PROTAC based on the small molecule inhibitor ibrutinib results in degradation not only of the wild type but also of the mutant protein, while ibrutinib targets only the wild type protein. P38 is involved in cell survival, migration and proliferation in cancer, and has 4 known isoforms. PROTACs composed of the same single inhibitor, the same E3 ligase and differing linker attachment sites provided isoform specific TPD, bypassing the need for complex isoform-specific inhibitor development. BCL-xL (B-cell lymphoma-extra-large) is overexpressed in many cancer types. The current BCL-xL inhibitors can also target platelets, resulting in severe side effects. Using the inhibitors in the context of a PROTAC, where the E3 ligase chosen is expressed at very low levels in platelets, like CBRN, can restrict the degradation of BCl-xL to cancer cells, and considerably improves patients' quality of life (1-4).
There are many other examples available in the literature, indicating the broad applications and potential of PROTACs for cancer therapy. Recently PROTAC refining strategies have been under investigation, leading to the development of PROTACs that work only under certain cellular conditions, such as phospho- and photo-PROTACs (1), and extend the specificity of this technology.
Finally, PROTACs can also be used in the treatment of viral infections. One example is in the treatment of Hepatitis C (HVC) where a PROTAC makes use of telaprevir (HCV protease inhibitor) to induce the degradation of the HVC NS3/4a protease (1). PROTACs targeting SARS-CoV-2 may reveal a useful therapeutic approach for patients severely affected.
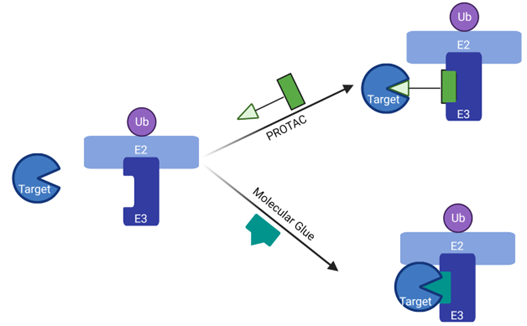
Figure 1: Schematic mode of action of PROTAC® and Molecular Glue.
PROTACs bring the Target into contact with the E3 Ligase via a POI-binding domain and an E3 Ligand connected through a linker. Molecular glues, on the other hand, simply stabilize the binding between target and E3, similarly to a glue.
Molecular Glues
Molecular glue (MG) degraders also bring together a POI and an E3 ligase, resulting in an outcome similar to PROTACs. However, they do not have a linker. Unlike PROTACs, MGs do not directly recruit E3 ligases to target proteins, but instead stabilize the interaction between the target protein and E3 ligase, resulting in enhanced ubiquitination of the target. MGs have a lower molecular weight and are more compact than PROTACs. Their design is more complex and this has limited the number of molecular glues developed so far. Examples include thalidomide and pomalidomide, which connect transcription factor IKZF1/3 to the E3 ligase CBRN (1).
Double-mechanism degraders can combine properties of PROTAC and molecular glues in one molecule. One example is GBD-9, that can act as PROTAC to degrade BTK and molecular glue for GSPT1 (G1 to S phase transition 1) by recruiting CRBN (1, 5).
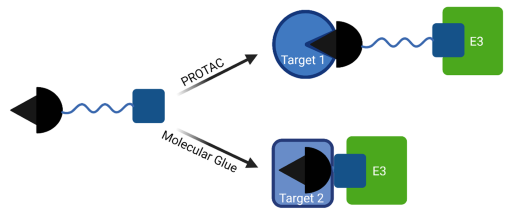
Figure 2: Schematic mode of action of double-mechanism degraders, such as GBP-9.
LyTACs
LyTACs expand the portfolio of targets by using lysosome degradation, extending the target pool to membrane and extracellular proteins. They are formed by a POI ligand, a linker and a lysosome targeting receptor (LTR) ligand. The formation of the tripartite complex with the POI and TRL results in endocytosis and finally degradation in the lysosome. The use of tissue specific LTRs allows LyTAC to be tissue specific, versus the use of inhibitors that can target all organs and result in significant side effects.
Although more recent than PROTACs, LyTAC have been applied in neurodegenerative, autoimmune diseases and cancer treatment. For example, conjugation of endocytosis-triggering poly-M6P (mannose-6-phosphate) to an EGRF antibody and poly-M6P with anti-PD-L1 antibody result in EGFR and PD-L1 degradation (1).
Several other methodologies targeting the lysosomal pathway have been developed and hold great promise for therapy (see Table).
| TPD Type | Molecular Composition | Mechanism | Example of POI |
|---|---|---|---|
| PROTAC (Proteolysis-Targeting Chimera) | POI-binding ligand Linker E3 Ligase ligand |
Target protein Ub and proteosomal degradation | ER, AR, BCL-xL, STAT3, EGFR, EZH2, BRAF, HPK1, FAK, BTK, NS3/4a |
| MG (Molecular Glue) | POI-binding ligand E3 Ligase ligand |
Target protein Ub and proteosomal degradation | IKZF1/3 |
| LyTAC (Lysosome-Targeting Chimera) | POI-binding ligand Linker LRT ligand |
Lysosome degradation via endocytosis | EGFR, PD-L1 |
| Bispecific Aptamer Chimera | POI-binding ligand DNA aptamer linker LRT ligand |
Lysosome degradation via endocytosis | PTK-7, MET |
| AbTAC (Antibody-based PROTAC) | Bispecific antibodies targeting POI and transmembrane E3 ligase | Lysosome degradation via endocytosis | |
| GlueTAC | Nanobodies binding antigen and cell penetrating peptide and lysosme sorting sequence (CPP-LSS) | Lysosome degradation via endocytosis | PD-L1 |
| AUTAC (Autophagy-Targeting Chimera) | cGMP-based degradation Tag Linker POI ligand |
Lysosome degradation via autophagy of cytoplasmic proteins and mitochondria | Mitochondria |
| ATTEC (Autophagosome Tethering Compound) | POI ligand LC3 ligand |
Lysosome degradation via autophagocytosis | Huntingtin |
| AUTOTAC (AUTOphagy-Targeting Chimera) | P62 ZZ domain POI binding domain |
Lysosome degradation via autophagy | Misfolded proteins such as Tau, AR |
| CMA-based degrader (Chaperone mediated autophagy (CMA)-based degrader) | Cell membrane penetration sequence POI binding domain CMA targeting motif |
Lysosome degradation via CMA | Mutant Huntingtin, PSD-95, DAPK1, α-synuclein |
Conclusion
The constant refinement of therapeutic approaches brings not only innovative strategies that can target previously “undruggable” proteins involved in disease, but also improvement in patient care. TPDs make use of the knowledge of the molecular pathways involved in proteostasis, such as the ubiquitin-proteasome and lysosomal degradation pathways, and disease. PROTACs, LyTACS and other TPD strategies will continue to expand our repertoire of targeted therapies and bring therapeutic benefits. BPS Bioscience can support your efforts in these fields with our innovative reagents and customer-tailored services.
References
(1) Zhao, L. et al, 2022 Signal Transduction and Targeted Therapy. 7 (113): https://doi.org/10.1038/s41392-022-00966-4
(2) Samarasinghe K. et al., 2021 Cell Chem Biol. 28 (7): 934-951.
(3) Ishida T., et al., 2021 SLAS Discov. 26 (4): 484-502

