Cellular PARylation: From Inquiry to Insight in Drug Discovery
Summary
Inhibitors of poly (ADP-ribose) polymerases (PARPs) have been game-changing in cancer therapy, demonstrating the potential efficacy of synthetic lethal drugs targeting complementary pathways within the DNA damage response network. However, the evaluation of poly-ADP-ribosylation in intact cells is difficult, slowing the development of candidate drugs. BPS Bioscience’s LysA™ Universal PARylation Assay Kit quantifies and compares amounts of PARylated proteins present in cellular extracts, allowing to determine compound efficacy in cells in culture, and bridging the gap between biochemical assays and animal testing. Here we discuss the principles of the assay and provide experimental examples of how to evaluate PARP or PARG inhibitors in intact cells.
Background
Addition of ADP-ribose units to proteins is a vital post-translational modification involved in cellular processes such as DNA repair, chromatin remodeling, and control of gene expression. It mainly happens in proteins linked to the DNA Damage Response (DDR). This addition comes in two flavors: mono-ADP-ribosylation (a single ADP-ribose unit is added, also termed MARylation) and poly-ADP-ribosylation (multiple units are added in linear or branched forms), also known as PARylation.
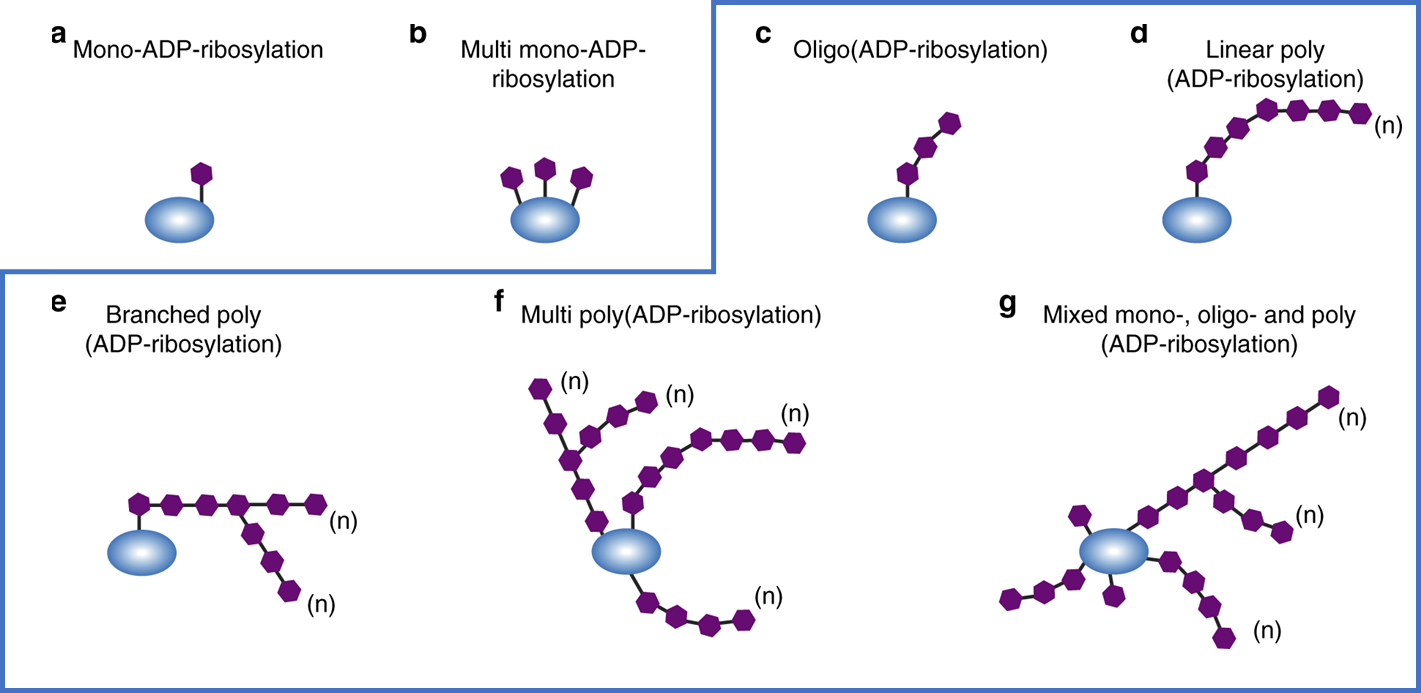
Figure 1: Illustration of various forms of mono and poly ADP-ribosylation. The anti-PAR antibody used in the assay described below recognizes various forms of poly-ADP-ribosylation as shown in the blue box. Figure used without modification, O’Sullivan et al, doi: 10.1038/s41467-019-08859-x, CC BY 4.0
PARylation is orchestrated by a family of 17 enzymes known as poly (ADP-ribose) polymerases (PARPs), with PARP1 to PARP5 catalyzing PARylation, and the other PARPs performing MARylation (1). PARP1, PARP2, and PARP3 have a strict DNA-dependency. PARP1 is the main player, since it is responsible for a whopping 80% of the total cellular PARylation (1). PARP1 and PARP2 are mostly involved in DNA repair, orchestrating the DDR network. PARP2 also controls epigenetics, proliferation, inflammation, and development (5, 6). In contrast, PARP1 alters transcription and induces apoptosis when DNA is damaged beyond repair. PARylation is reversible and can be undone by PAR erasers, such as poly (ADP-ribose) glycohydrolase (PARG) (2, 3).
PARP1 and PARP2 sense single-strand DNA breaks, latch onto the damaged area, then add PAR chains to their own backbone, to histones, and to other repair proteins in order to recruit and activate these proteins. PARylated PARP1 or PARP2 then detach from the DNA due to the allosteric hindrance of negatively charged PAR chains, allowing other proteins to access the DNA and initiate the repair process. PARG's job is to remove the PAR chains, recycling the repair proteins to their inactive form, a crucial step in various DNA repair processes (4).
PARP1/2 inhibitors have been successful drugs for cancer treatment, leading to synthetic lethality when homologous recombination repair (HRR) is defective in the tumor cells (7). Indeed, there are now four FDA-approved drugs in clinical use in the US, with many more in the pipeline.
On the other hand, dysregulation of PARG activity is linked to various diseases, including cancer (8). Targeting PARG for therapy is gaining traction as a strategy for sensitizing cancer cells to DNA damage-inducing agents (9). Blocking PARG activity leads to PAR buildup on proteins and interferes with DNA repair mechanisms, ultimately leading to cell death. PARG inhibitors, therefore, are a promising therapeutic avenue, especially when combined with DNA damage-inducing radiation or chemotherapy. Apart from PARG, other PAR erasers contribute to PAR turnover and DNA repair, like MacroD1 and MacroD2 (macrodomain-containing proteins), TARG1 (also known as ARH3), and NUDT16.
Why measure cellular PARylation?
Cell-based assays are an essential part of the drug development process, as they allow researchers to evaluate the biological activity of drug candidates in living cells and gain a better understanding of their mechanism of action. Cell assays are necessary to evaluate the compounds’ membrane permeability issues and interferences within a complex system, and overall provide a more accurate representation of the complex interactions between compounds and living cells, as compared to biochemical assays.
Thus, adding a drug candidate such as PARG or PARP inhibitors to a cell of interest and measuring the resulting levels of PARylation can provide precious information on the effectiveness of the compound in intact cells. The LysA™ Universal PARylation Assay Kit evaluates PARylation in cellular extracts and allows determination of compound IC50, comparison in the IC50 of various compounds, the effect of a compound in cells with specific DDR defects, and more.
Example applications
- Measure total PAR levels after cell stress induced by chemotherapy drug (indicates, for example, if PARP was activated and to what extent).
- Screen or determine the IC50 of inhibitors of the PARP family (PAR writers).
- Screen or determine the IC50 of inhibitors of the PARG family (PAR erasers).
- Validate a new cellular model for the study of PAR homeostasis.
- Evaluate the possible synergistic effect of treatment combinations.
- Quantify PAR levels in biological samples using a defined PAR standard.
Principle of the assay
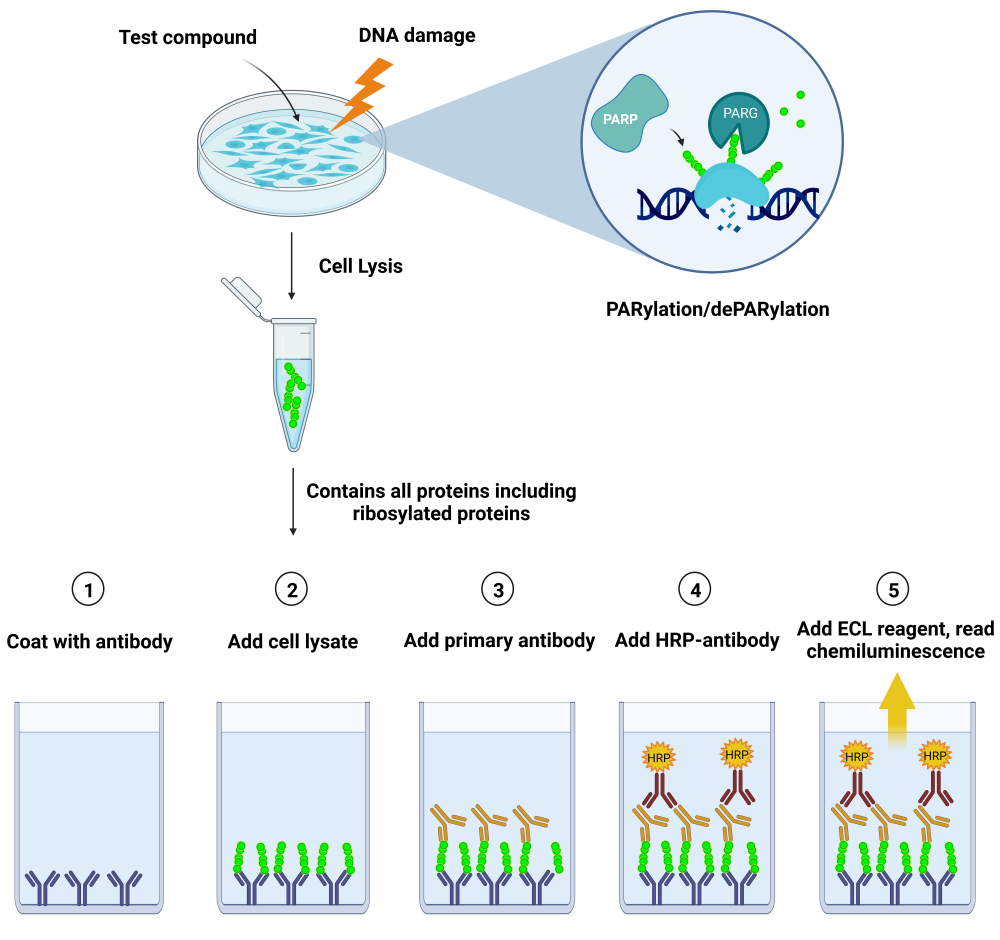
The LysA™ Universal PARylation Assay Kit is a sandwich ELISA-based assay designed to analyze the level of total PARylation present in cellular extracts. The kit includes a PAR standard for absolute quantification. The assay detects differences in protein PARylation levels resulting, for example, from inducing the DNA damage response or from exposure to PARP inhibitors or to PARG inhibitors.
Principle: a 96-well plate is coated with an anti-PAR antibody specific for PARylated chains. Lysates from cells are added to the coated wells, and PARylated proteins present in the cell lysates are captured by the antibody. This is followed by an incubation with an anti-PAR detection antibody, then a secondary HRP-conjugated antibody. Addition of a chemiluminescent HRP substrate provides a luminescence signal that directly correlates with the level of cellular PARylation.
The assay relies on a specific anti-PAR antibody and does not detect mono-ADP-ribosylation (MARylation).
Keeping in mind that PAR levels reflect the homeostasis between PAR writers and PAR erasers, it is expected that adding specific inhibitors and/or using DNA-damaging agents will affect this balance. For example, adding PARG inhibitors pushes the system toward increased PARylation by blocking PAR removal, whereas adding PARP inhibitors decreases protein PARylation.
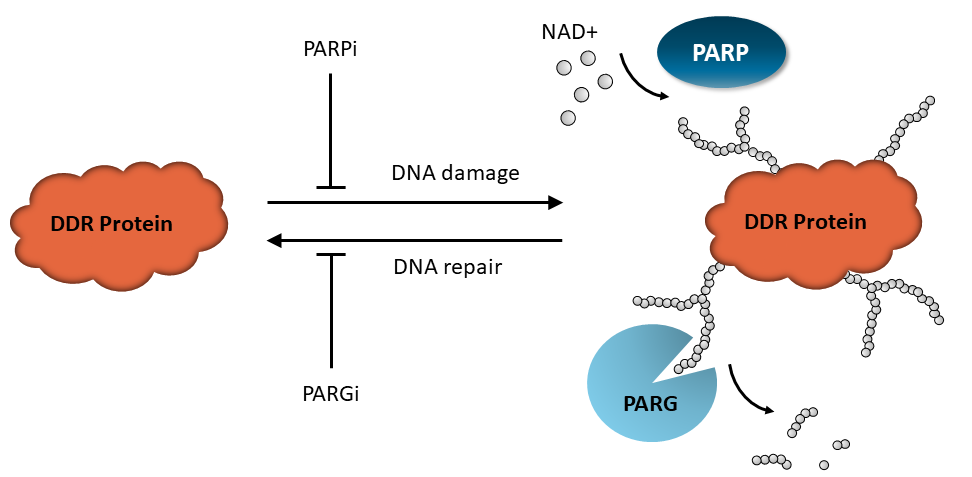
Figure 3: PARylation homeostasis. DNA damage activates PARP proteins, leading to the formation of PARylated DNA repair proteins, including PARP themselves. Once DNA repair has been initiated, PAR erasers such as PARG remove the ADP-ribose units from the proteins, recycling them to respond to DNA damage again. Either reaction can be blocked by specific inhibitors, resulting in the accumulation of PARylated proteins or in the prominence of the non-PARylated form, depending on context.
Examples of cellular experiments analyzed using LysA™ Universal PARylation Assay Kit
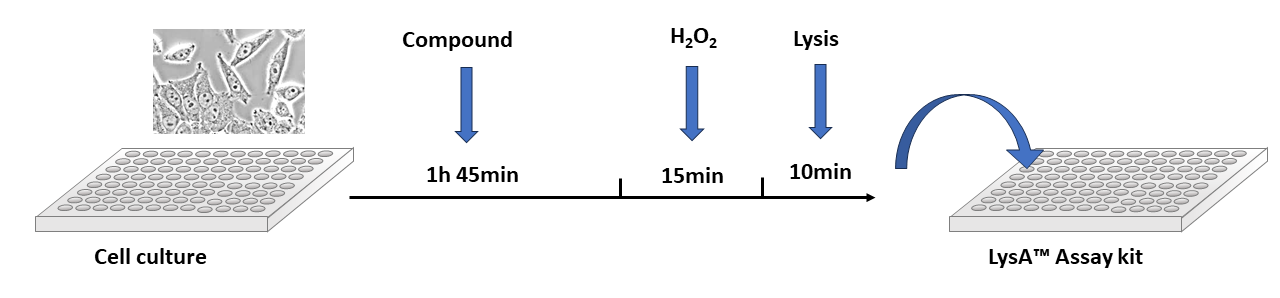
Figure 4: Cells in culture are treated with a compound of interest before DNA damage is induced. Cells are lysed and analyzed using LysA™ Universal PARylation Assay Kit.
1) Titration of the PARG inhibitor PDD00017273 in various cell lines
- HEK293, MCF7, MDA-MB-231 and HeLa cells were seeded in 96-well cell culture plates at a cell density of 0.5 x 105 cells/well and were cultured in their corresponding growth media until they reached approximately 70% confluency, approximately 24 hours for HeLa and HEK293 cells and 48 hours for MCF7 and MDA-MB-231 cells.
- Without changing the growth medium, cells were treated without or with increasing concentrations of PARG inhibitor PDD00017273 (MedChem Express #HY-108360) for 1 hour and 45 minutes at 37°C prior to adding hydrogen peroxide (H2O2, 500 µM final) for an additional 15 minutes to induce DNA damage. Note that PDD00017273 stock solution was initially prepared in DMSO, and the concentration of DMSO (1%) was kept constant across all conditions including the no-inhibitor control.
- After treatment the medium was gently removed from the plates and the cells were washed once with ice-cold PBS (Phosphate Buffer saline), placed on ice, and lysed using 50 µl/well of Modified RIPA Lysis Buffer (BPS Bioscience #82126) supplemented with protease inhibitor cocktail and ADP-Ribosylation Cycle Inhibitor Mix (BPS Bioscience #82130).
- After 10 minutes on ice, lysates were pipetted up and down several times and collected.
- PARylation levels were analyzed immediately using the LysA™ Universal PARylation Assay Kit. The entire volume of each lysate (50 µl/well) collected in the previous step was loaded to the corresponding well in a white 96-well module Maxisorp™ plate coated the day before the experiment.
- After following the LysA™ Universal PARylation Assay Kit protocol (BPS Bioscience #82123), luminescence was measured using a Bio-Tek microplate reader.
- Results are expressed as percent of total PARylation (in which the maximum PARylation level was set at 100%).
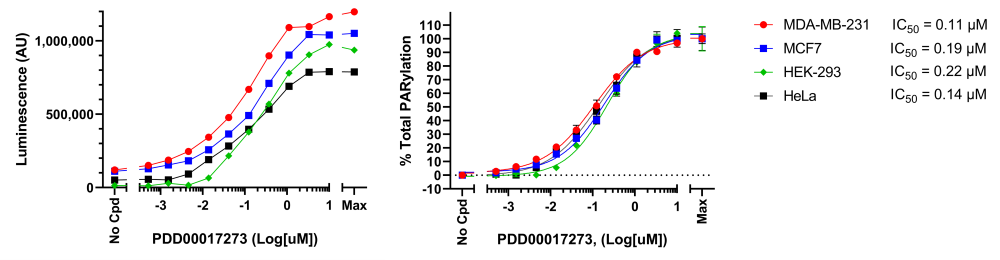
Figure 5: Effect of PARG inhibitor PDD00017273 on H2O2-induced protein PARylation in various cell lines.
Cells were treated with increasing concentrations of PARG inhibitor and H2O2 as described above. Cell lysates were analyzed using LysA™ Universal PARylation Assay Kit. Results are expressed as raw luminescence signal (left) or normalized as percent of total PARylation in which the maximum PARylation for each cell line was set to 100% (right). For each cell line, the IC50 was calculated and plotted using GraphPad Prism software. Data represents biological duplicates from a single experiment.
2) Titration of several PARP inhibitors in HEK293 cells
- HEK293 cells were seeded in 96-well cell culture plates at a density of 0.5 x 105 cells/well and were cultured in their normal growth medium until they reached a confluency of about 70%, approximately 24 hours.
- Without changing the growth medium, cells were treated with 10 µM PARG inhibitor PDD00017273 (MedChem Express #HY-108360) to prevent de-ribosylation, in combination with increasing concentrations of PARP inhibitors Olaparib, Talazoparib, and AZD5305 (set of PARP inhibitors, BPS Bioscience #78318) for 1 hour and 45 minutes at 37°C. Hydrogen peroxide (H2O2, 500 µM final) was then added for an additional 15 minutes to induce DNA damage. Note that the inhibitor stock solutions were initially prepared in DMSO, and the concentration of DMSO and PARGi was kept constant (1% and 10 µM, respectively) across all conditions including the control without PARP inhibitor.
- The medium was gently removed from the plates and the cells were washed once with ice-cold PBS, placed on ice, and lysed using 50 µl/well of Modified RIPA Lysis Buffer (BPS Bioscience #82126) supplemented with protease inhibitor cocktail and ADP-Ribosylation Cycle Inhibitor Mix (BPS Bioscience #82130).
- After 10 minutes on ice, lysates were pipetted up and down several times and collected.
- PARylation levels were analyzed immediately using the LysA™ Universal PARylation Assay Kit. The entire volume of each lysate (50 µl/well) was collected in the previous step was loaded to the corresponding well in a white 96-well module Maxisorp™ plate coated the day before the experiment.
- After following the LysA™ Universal PARylation Assay Kit protocol (BPS Bioscience #82123), luminescence was measured using a Bio-Tek microplate reader.
- Results are expressed as percent of total PARylation (in which the PARylation level measured in absence of inhibitor was set at 100%).
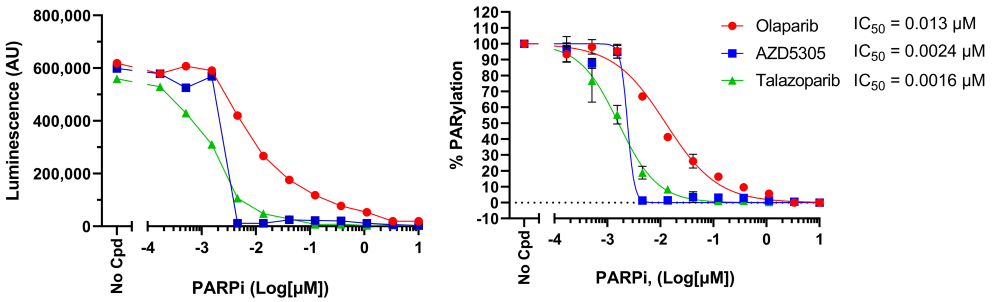
Figure 6: Effect of PARP inhibitors on H2O2-induced protein PARylation in HEK293 cells.
Cells were treated with increasing concentrations of PARP inhibitors and H2O2 in the presence of PARG inhibitor, as described above, and cell lysates were analyzed using LysA™ Universal PARylation Assay Kit. Results are expressed as raw luminescence signal (left) and normalized as percent of total PARylation in which maximum PARylation was set to 100% (right). For each condition, the IC50 was calculated and plotted using GraphPad Prism software. Data represents biological duplicates from a single experiment.
Conclusion
Data show the usefulness of measuring cellular PARylation levels to evaluate the efficacy of PARP or PARG inhibitors. BPS Bioscience’s LysA™ Universal PARylation Assay Kit provides a convenient, simple assay to compare the effect of various compounds in intact cells.
References
(1) Ummarino S, et al. 2021 Genes 12(3): 446.
(2) O'Sullivan J, et al. 2019 Nat Communications 10(1): 1182.
(3) Harrision D, et al. 2020 Front Mol Biosci. 7: 191.
(4) Murai J, et al. 2012. Cancer Res. 72(21): 5588-99.
(5) Ali SO, et al. 2016 Am. J. Cancer Res. 6(9): 1842-1863.
(6) Rose M, et al. 2020 Front. Cell Dev. Biol. 8: 564601.
(7) Rudolph J, et al. 2022 PNAS USA, 119(11): e2121979119.
(8) Marques M, et al. 2019, Oncogene. 38(12): 2177-2191.
(9) James DI, et al. 2016, ACS Chem Biol. 11(11): 3179-3190.

