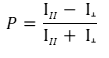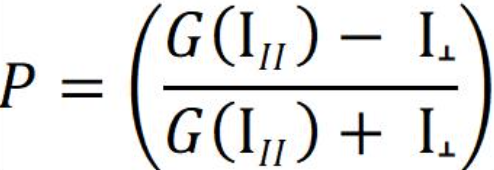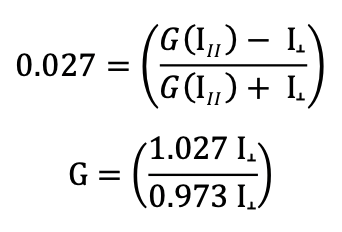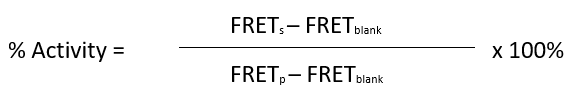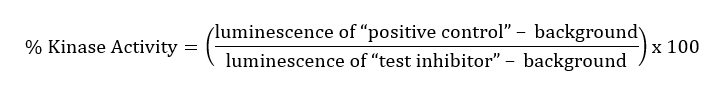How do I thaw recombinant proteins?
Thaw on ice and gently mix prior to use. DO NOT VORTEX. Perform a quick spin before opening the tube. Avoid multiple freeze/thaw cycles. Enzymes are particularly sensitive to freeze/thaw and may lose some activity if thawed again.
When proteins are diluted to perform an assay do not freeze and re-use the diluted protein.
What to do if I need to freeze the protein again?
If the protein is going to be used more than once, aliquot into small volumes upon first thaw, and flash freeze for long term storage. To avoid evaporation or stability issues, aliquots should not be less than 5 µl in volume. Do not thaw again more than once. Perform a quick spin before opening the tube to recover the full content of the tube.
Where do I find information on the concentration of the protein?
The exact concentration of a protein is lot-specific and is indicated on the vial containing the protein.
Why is there a FLAG peptide in the formulation of my protein?
Recombinant proteins containing a FLAG-tag (a FLAG-tag consist of amino acids DYKDDDDK) may be affinity-purified using anti-FLAG antibody beads and eluted with free FLAG peptide. The FLAG peptide contained in the protein formulation buffer is just a left-over from the purification process. It is not due to cleavage of the tag from the protein but is residual from the elution step.
The free FLAG peptide will not interfere with most types of assays. However, if desired, it may be removed from the final formulation by monoQ upon request (a fee may be added).
Can you help me convert a mass concentration to a molar concentration for purified proteins?
Use formula: [µM] = [µg/mL] / MW(kDa)
Example: If you receive 200 µg of a 40 kDa protein in 100 µl, then the mass concentration is 2,000 µg/ml (2mg/ml). Therefore, your molar concentration is 2,000/40 = 50 µM (0.05 mM). Make sure to observe all the units of measurement.
What is the proper storage condition for recombinant proteins?
Store at –80°C. Thaw on ice, spin briefly in a centrifuge so all liquid is at the bottom of the tube, aliquot ≤ 5 µl to individual single-use tubes and re-freeze immediately. Avoid repeated thaw/freeze cycles. Diluting proteins prior to storage is not recommended and may require the addition of a carrier protein such as BSA (bovine serum albumin; 0.1%-0.5%). If dilution is necessary, keep the final protein concentration above 10 µg/ml.
Why is the molecular weight (MW) in SDS-PAGE different from the expected MW?
The expected MW of a recombinant protein is calculated from its amino acid sequence. Most commonly, post-translational modifications may change the apparent MW of the recombinant protein in SDS-PAGE, for example when the protein is expressed in a system that allows for glycosylation. In these cases, a broadening of the band may also be seen due to the addition of heterogeneous complex glycans in expression systems such as HEK293.
What is the difference between a protein expressed in E. coli, Sf9 baculovirus, or mammalian cells?
Bacterially expressed proteins have high expression rates with minimal post-translational modifications. Examples of post-translational modifications not present in bacterial recombinant proteins include glycosylation, tyrosine phosphorylation and epigenetic post-translational modifications. Despite a limited protein folding machinery, bacterial expression is preferred when proteins are properly folded and post-translational modifications are not needed. Insect cells are eukaryotic, therefore the recombinant proteins expressed in this system often have the same or very similar post-translational modifications as observed in mammalian cells. One major difference between insect cells and mammalian is that proteins expressed in insect cells contain the simple glycan (GlcNAc)2Man9, added through N-glycosylation, whereas proteins expressed in mammalian cells contain more complex and heterogeneous glycans.
How do you purify recombinant proteins?
Usually, our recombinant proteins contain a tag to facilitate their detection and purification. Proteins are affinity-purified according to their tag, for example using Ni-NTA resins for a 6xHis tag, protein A columns for a Fc tag, and so forth. Gel filtration may be used to assess aggregation.
What is HiP™?
HiP™ proteins, or High Purity Proteins, are affinity-purified recombinant proteins with a purity level >90% AND less than 10% aggregation as assessed by gel filtration. Our HiP™ trademark designates our highest standard of quality for recombinant proteins.
What kind of His-tag does BPS use?
His-tags are typically composed of 6–10 consecutive histidine residues at either end of the protein of interest. The protein description on our website usually indicates where the tag is located, and whether it is a 6xHis or a 10xHis. BPS commonly uses 6xHis-tag, which has a MW of 0.8 kDa, however 9xHis or 10xHis may be added as custom tags. His-tags facilitate protein detection or immunoprecipitation of the protein using an anti-His tag antibody, or purification using immobilized metal affinity chromatography (IMAC), the most common being Ni-resin. The tag's small size reduces interference with protein structure or function.
What is an Avi-Tag™?
The Avi-Tag™ is an optimized 15-amino acid peptide used as substrate by biotin protein ligase BirA. It allows the highly selective addition of one biotin molecule to the lysine residue of the tag by enzymatic biotinylation. It can be added to either end of a protein and used for detection, immunoprecipitation or purification. Avi-Tag™ biotinylation is more precise than other biotinylation methods, as the biotin moiety is added only to the Avi-Tag sequence. The molecular weight of the Avi-Tag™ is 1810 Da.
How are proteins biotinylated?
BPS uses the AviTag™ technology for most biotinylated products. Recombinant proteins are usually biotinylated in vitro using BirA ligase to covalently append biotin to the AviTag™, placed either at the N-terminal or the C-terminal end of the target. This method results in very good rates of biotinylation (typically ≥90%). To learn more, see our
BirA tech note.
What is a PreScission™ sequence?
A PreScission™ sequence is a short linker sequence that can be inserted between a protein of interest and a tag, and is used to remove the tag from the recombinant protein when desired. This is achieved using the PreScission™ protease, also known as the human rhinovirus (HRV) 3C protease. This protease specifically cleaves the amino sequences Leu-Phe-Gln↓Gly-Pro between the Gln and Gly residues.
What is an Fc tag?
The Fc (fragment crystallizable) region of an antibody is the C-terminal domain responsible for interaction with cell surface Fc receptors and with proteins of the complement system. The discovery that bacterial protein A and protein G bind strongly to the Fc portion of IgGs led to the widespread adoption of purification and precipitation methods using Protein A/G resins. Recombinant proteins in which the C-terminus has been fused to the Fc region of an IgG can be detected with anti-IgG antibodies, and can be immunoprecipitated or purified with protein A/G/L resins in a straightforward fashion.
I could not find my protein in your catalog. What now?
BPS Bioscience adds new proteins on a weekly basis. If you do not find your favorite protein, let us know. We are happy to
design new products upon request and modify existing proteins according to your specific needs.
What is the difference between KRAS isoforms A and B?
The KRAS (Kirsten rat sarcoma virus) gene is subject to alternative splicing, resulting in two isoforms: KRAS-A and KRAS-B. These isoforms differ by amino acids 151, 153, 165, and 166 and within the hypervariable region (amino acids 167–189). KRAS-B contains a long polybasic stretch, while KRAS-A has a shorter polybasic region with a palmitoylation site. These differences confer distinct biological characteristics to the two isoforms. When studying a mutant of KRAS, it is important to know which isoform is being studied to make sure that the correct wild-type isoform is used for comparison.
How should I store the cells?
Cells are shipped in dry ice, and upon receipt should immediately be thawed for culture or stored in liquid nitrogen. Do not keep the cells in dry ice for an extended time. Do not use a -80°C freezer for long term storage. Cells can be stored in liquid nitrogen for many years. For very long-term storage, it is recommended to thaw cells after about 5 years, expand them again and freeze new vials.
Contact technical support at
[email protected] if the cells are not frozen or in dry ice upon arrival.
How many cells does each vial contain?
Each vial of growing cells shipped from BPS Bioscience typically contains 2 million cells, unless specifically indicated on the website and the product datasheet. There may be slightly more than 2 million cells in a vial to ensure fast regrowth of the cell culture.
For growth arrested cells (used in our cell-based assay kits), the exact number of cells is usually not disclosed. Each vial provides a large number of cells that has been optimized for the specific cell-type and assay in consideration, so the assay can be performed immediately.
Do you sell inhibitors or agonists to use as internal controls in cell-based assays?
How do you perform cell lysis when using ONE-Step™ Luciferase Assay System?
The ONE-Step™ Luciferase Assay System consists of components A and B, which are mixed prior to adding the reagent to the cells. The reagent contains a lysis buffer, so there is no need to perform cell lysis separately. Just add the ONE-STEP™ reagent, wait for about 15 minutes, and read luminescence.
What are growth-arrested cells?
Growth arrested cells are engineered recombinant cell lines designed for single use. These cells are alive and suitable for one cell-based assay, but the cells cannot replicate and therefore do not proliferate in culture.
What type of Quality Control does BPS Bioscience perform before shipping the cells?
All cell lines are tested for bacterial and mycoplasma contamination prior to shipment. They are also tested for cell growth and viability at 24 and 48 hours after thawing. The corresponding Certificate of Analysis will be sent with the package containing the cells.
Why does BPS Bioscience supply two vials of cells?
Sometimes things do not go as planned. Cell viability may be compromised due to improper shipping, storage, or thawing conditions. The 10% DMSO (Dimethyl Sulfoxide) contained in the freezing medium is toxic to cells and should be removed immediately upon thawing. Be sure to minimize the time cells are in the freezing medium when thawing. Please read the thawing protocol provided in the datasheet.
Optimal cell culture media and growth conditions are also specified in the datasheet and on our website. The thawing process is very stressful to the cells, so the culture medium used for thawing and during the first few days of culture should not contain selection antibiotics (such as geneticin, hygromycin or puromycin).
If cells thawed from the first vial do not grow as expected, contact us at
[email protected] and we will troubleshoot with you before you thaw the second vial. Of note, it is recommended to expand the newly grown cells and freeze at least 10 vials at an early passage for future use.
Why do your growth media contain selection antibiotics?
Our recombinant cell lines have been engineered to express one or several genes following stable transfection of the cells. Briefly, cells are transfected with the gene of interest following standard procedures. A few days later, cells in which the gene of interest has integrated into the genome are selected using antibiotic pressure (geneticin, puromycin and hygromycin are the most common antibiotics used to generate our cell lines). Antibiotic pressure must be maintained during long term culture to avoid loss of the transfected gene.
How stable are your recombinant cell lines?
Cell lines are guaranteed to be stable for up to 15 passages when grown in our media and using the appropriate selection antibiotics. In our hands they are stable for at least 20 passages most of the time, however we recommend starting over from an early-passage frozen vial at passage 15 to ensure reproducibility of results.
Can I use cell culture media from other suppliers?
For best results, it is highly recommended to use optimized media from BPS Bioscience, which have been validated for each recombinant cell line. Other preparations or formulations of media may result in suboptimal performance. The
composition of each thaw medium and growth medium is specified on our website. Keep in mind that the lot-to-lot variation and provenance of FBS (Fetal Bovine serum) may noticeably influence cell growth parameters.
The cell culture media I purchased need to be stored at 4°C, but they arrived frozen on dry ice. Can I still use them?
To decrease packing and shipping costs for our customers, thaw and growth media or other components may be sent in the same package as the cells, in dry ice, and will be received frozen. This also ensures that media are received at the same time as the cells, as they are required for thawing and growing the cells. The media are stable when frozen and can be transferred to the recommended storage temperature (usually 4°C) without loss in activity.
What is the difference between a cell pool and a cell line?
A cell pool is obtained following antibiotic selection of a genetically modified cell population, without cloning. Thus, the cell pool contains all the cells that survived the antibiotic upon stable integration of the resistance gene. The resulting population may be somewhat heterogenous, however it better reflects the original cell population. Conversely, a cell line results from the cloning of antibiotic-selected cells using, for example, the limiting dilution method. Since a cell line originates from a single clone, it is homogeneous in terms of genetic modification or protein expression, however it may not reflect the original cell population as closely as a cell pool.
What kind of assay plate should be used to perform a cell-based assay?
Cells that grow attached do not grow on regular plastic, therefore cells should always be plated on a cell culture-treated plate. It is preferable to use a clear-bottom plate to be able to visually observe the cells under a microscope after plating, before the experiment, and at the end of the experiment if necessary.
- Colorimetric assay: regular clear plastic cell culture plate.
- Luminescence assay: clear-bottom, white assay plate. White plates are used so that the light created by the luciferase in a well will be reflected up or down to the reader component and not be dispersed to neighboring wells through a clear wall.
- Fluorescence assay: clear-bottom, black assay plate. Black plates are used so that the fluorescence signal in a well will be reflected up or down to the plate to the reader component and not be dispersed to neighboring wells through a clear wall.
How does BPS Bioscience determine the concentration of antibiotic to be used to maintain a genetically modified cell line?
The optimal concentration of each antibiotic is initially determined for each cell line by performing a kill curve.
What is a kill-curve?
A kill-curve is a dose-response experiment in which cells are cultivated in the presence of increasing concentrations of a specific antibiotic for a period of one week to ten days. The minimum antibiotic concentration that is both required and sufficient to kill all the cells is then used for antibiotic selection, and for maintenance of the genetically modified cell line.
Protocol:
Plate the cells in a 24-well plate in their respective complete growth medium. Plate a number of cells that allows cells to reach approximately 30-50% confluency a day later. The day after plating, add increasing concentrations of the antibiotic of interest. Include a medium-only (no antibiotic) control. It is recommended to perform the dose-response in duplicates.
The useful range of concentration depends on the antibiotic.
For example:
G418 0.1 to 2.0 mg/ml
Hygromycin 100 to 500 µg/ml
Puromycin 0.25 to 10 µg/ml
Replace the cell culture medium, maintaining the antibiotic concentration, every 3-4 days for up to 10 days. Examine the cells under a microscope every day for signs of cell death. Make note of the antibiotic concentration that is necessary and sufficient to kill all cells after 10 days.
At what percentage of CO2 should I set my cell culture incubator?
The CO2 should be set at 5%.
CO2 present in the air (atmospheric) and dissolved in the cell culture medium affects the pH of the medium. This can be seen by changes in the color of media containing phenol red, which is a pH indicator: as pH decreases and the medium becomes more acidic, the phenol red turns orange, then yellow. Conversely, as pH increases and the medium becomes more basic, the phenol red turns pink.
Increasing the CO2 level decreases the pH of the medium and vice-versa. Atmospheric CO2 is regulated by the incubator and therefore remains constant. However, cell metabolism produces CO2, which is released in the medium as cells grow and decreases the pH of the medium. To stabilize the pH in the presence of higher levels of CO2, sodium bicarbonate NaHCO3 is used as a buffer. The concentration of sodium bicarbonate added to the medium is calculated to counteract specific amounts of CO2.
Media containing 1.5 to 2.2 g/L sodium bicarbonate are used to grow cells in 5% CO2. Media containing 3.7 g/L sodium bicarbonate are used to grow cells in 10% CO2. BPS Bioscience uses media containing the lower range of sodium bicarbonate, therefore cells grown in our media need to be maintained in a 5% CO2 incubator.
How can I verify the health of my cells?
Trypan blue staining is routinely performed to verify the viability of cells in suspension. It is used for the colorimetric detection of dead cells.
Trypan blue is a highly charged molecule that does not pass through the cell plasma membrane. However, the dye will penetrate cells with a damaged plasma membrane. Therefore, damaged or dead cells appear blue while healthy cells exclude the dye and remain unstained. The method requires observation under the microscope with a manual count, or an automated cell counter with colorimetric detection capability.
Note that trypan blue staining is a very basic marker of cellular heath and does not provide mechanistic information on cell death. In addition, the dye quenches fluorescence and will interfere with fluorescence staining.
Do you sell inhibitors or agonists to use as internal controls?
Yes, we do sell support reagents such as
inhibitors and agonists. Note that an internal control inhibitor or agonist may be included with some assay kits. Check in the table of “Assay Kit Components” of the product page.
What is the composition of your buffers and reagents?
In some cases, the composition of a reagent will be available from the web page and the technical datasheet corresponding to the product of interest. However, the composition of many of our reagents is proprietary. If you need to know whether a specific chemical is present in one of our proprietary reagents (for example, dithiothreitol), contact us with your question at
[email protected].
What is a homogeneous assay?
An assay in which no wash steps are required to remove unbound molecules, since the presence of unbound molecules does not interfere with the reading. This reduces the number of steps necessary to perform the assay and makes it more amenable to high-throughput applications. Fluorescence Polarization, TR-FRET, AlphaScreen™, and Fluorogenic assays are typically homogeneous assays. ELISAs (Enzyme-linked immunosorbent assays) are heterogeneous assays with several wash steps required throughout.
Can I use cell lysates or other complex extracts with the kit?
No. Our biochemical assay kits are based on the validated activity of a recombinant, affinity-purified protein of interest. They are designed to directly test the effect of compounds on the target enzymatic activity, or on the binding between two partners. They are not designed to assay enzymatic activity present in a complex system such as cell lysates and we do not recommend it as it would not be possible to assess the specificity of the signal generated.
How do I store the kit components?
Store each kit component as indicated in the “Supplied Materials” table displayed in the datasheet or the “Format” table shown on the product page.
Most kits contain purified recombinant proteins that are very sensitive to temperature and to freeze/thaw cycles. Kits are shipped with dry ice to maintain the full activity of the kit components. Contact technical support at
[email protected] if you notice an issue with the shipment.
Where can I find the protocol of the assay kit?
Detailed assay protocols are provided in the datasheet corresponding to the assay kit. Datasheets are available on our website and can be accessed from the product page.
Always read the protocol carefully from start-to-finish before starting an experiment.
How do I know if BPS Bioscience has tested a reference compound in an assay?
The assay datasheets usually show all the results validated by our team of scientists and the protocols used to generate the data. Additional information about other researchers’ use of an assay may be found in publications citing our assay (look in the Citations section of the product page).
I spilled a reagent used in my assay kit. Can I order it separately?
Yes, some reagents are available separately. Contact your Sales Representative or send us a message providing the name and catalog number of the reagent at
[email protected].
Do you sell inhibitors or agonists to use as an internal control in biochemical assays?
We often provide an internal control with our assay kit. In addition, we sell support reagents such as
inhibitors and agonists.
Can I use the kit more than once?
If the assay plate is going to be used more than once, prepare enough reagents for this portion of the assay and aliquot the remaining undiluted reagents into single-use aliquots depending on how many times the assay plate will be used. Store the aliquots at -80°C or at -20°C as appropriate.
Keep in mind that some enzymatic activity will be lost with each freeze/thaw cycle.
What precautions should I take using the proteins supplied with the kit?
The concentration of each protein is lot-specific and is indicated on the tube. Always verify the stock concentration before using the protein.
Thaw on ice and gently mix prior to use. DO NOT VORTEX. Perform a quick spin before opening to recover the full contents of the tube. Proteins should be kept on ice until use. If dilution is required, use the Assay Buffer provided with the kit. Discard unused diluted protein at the end of the experiment.
What is a stepwise dilution?
A stepwise dilution is used when the stock solution of a component, such as a protein, is provided at high concentration. It is especially recommended in instances when diluting the reagent in a single step would result in a very large volume and/or use most of the buffer.
For example, if the protocol requires diluting a reagent 1,000-fold in Assay Buffer, do NOT dilute 10 µl of reagent in 10 ml of assay buffer. Instead, dilute 10-fold by adding 10 µl of reagent to 90 µl of Assay Buffer, then dilute again 100-fold by adding 10 µl of the previous dilution to 990 µl of Assay Buffer.
How do I dissolve the test compound that I will use in the assay?
It depends on the compound. If it is water-soluble, dissolve in the Assay Buffer as recommended in the protocol and prepare serial dilutions in the Assay Buffer. Controls should use the Assay buffer as well.
If the compound is soluble in organic solvents, such as DMSO, dissolve in 100% DMSO (or other appropriate solvent) at a high concentration, then dilute in Assay Buffer. Keep in mind that if you are testing a serial dilution, the concentration of DMSO should be kept constant across the dilution and controls.
Note that most of our assays are validated using DMSO as an organic solvent. Validated experimental conditions are indicated in the datasheet. We recommend that you read the protocol carefully from start-to-finish before starting the experiment.
Could some common chemicals affect my assay?
Some assays are sensitive to the presence of a particular chemical. Examples of chemicals or buffer components known to alter the performance of biochemical assays: DMSO >1%, strong acids or bases, ionic detergents, high concentrations of salt. When our scientists have determined that a chemical or experimental condition has unwanted consequences in an assay, it will be indicated on the datasheet.
What kind of plate should I use?
Assay kits include a 96-well or a 384-well plate to perform the experiment. Low-binding plates are used with homogeneous assays such as TR-FRET assays. White plates are used with luminescent or chemiluminescent assays, whereas black plates are used with fluorogenic assays.
Some ELISA plates may be pre-coated, such as nickel-coated plates to capture His-tagged proteins or Avidin-coated plates to capture biotinylated proteins (NeutrAvidin™ Plate
#79512). Be sure to store the plates as directed in the datasheet.
What is an IC50?
IC50 (Inhibitor Concentration 50) is a quantitative measure of the potency of a compound in inhibiting a biological or enzymatic function, in vitro. Specifically, it measures the concentration at which the compound inhibits a given biological or enzymatic process by 50%.
What is the difference between IC50 and EC50, and when do I use one or the other?
IC50 and EC50 are used in dose-response curves to determine the potency or effectiveness of a substance. The lower the value the greater the potency as it means that lower concentrations of the substance achieve the 50% effect.
IC50 (Inhibitory Concentration 50) represents the concentration of compound required to inhibit an effect by 50%. Therefore, it is used in studies involving inhibitors or neutralizing peptides and antibodies. For instance, in drug development, IC50 values are used to assess the potency of inhibitors against enzymes or to neutralize ligand/receptor binding.
EC50 (Effective Concentration 50) represents the concentration of compound required to produce 50% of a desired effect (e.g., 50% of the maximum achievable response). It can be used to measure the potency of agonists such as a receptor ligand, hormone, or neurotransmitter. It is also used in pharmacology studies to determine the potency of a drug in eliciting a therapeutic effect.
How do you determine the IC50 of a compound?
It is determined by performing a dose-response analysis in which increasing concentrations of the compound are added to the cells (biological effect) or to the purified enzyme (biochemical effect) and the desired effect is quantified. The dose-response must be designed to capture potency, with lower concentration(s) showing lack of effect and high concentration(s) showing maximum effect.
-
Design a serial dilution of the compound of interest in 3-fold increments. BPS Bioscience uses 9 to 10 concentrations in a dose-response.
-
Include internal controls appropriate for the type of assay, and a control without compound. All assays require a “blank” (in the absence of cells or enzyme, signal originating from the reagents).
-
Confirm that you are looking at the active enzyme or biological effect. In some cases, you may need to add an agonist to activate the cells or the enzyme.
Why does BPS Bioscience recommend 3-fold dilutions to measure inhibitors IC50?
Most of the time, BPS Bioscience uses a base 10 Log scale for graphs, in which 3-fold dilutions are logarithmic and evenly spaced, whereas linear dilutions on a log scale may be clumped together. See these two theoretical plots: on the left, the compound concentrations were selected linearly and appear clumped by two. On the right, the compound concentrations were selected logarithmically and are evenly spaced. The logarithmic scale provides better sampling and is a better experimental design.
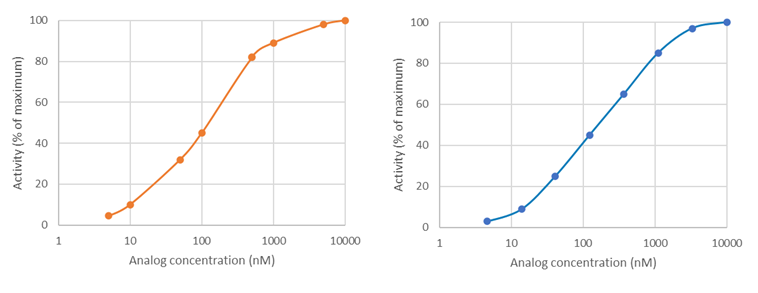
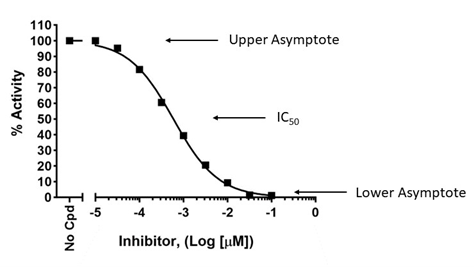
How do you know if you did not choose the correct concentration range?
To accurately determine an IC
50, a dose-response must be designed to capture true potency, with lower concentration(s) showing lack of effect and high concentration(s) showing maximum effect. This allows to determine the higher and lower asymptotes on the graph, which are then used to calculate the position of the 50% value, as shown below (upper graph). If the higher concentrations do not cross the x-axis and do not reach a plateau, the true IC
50 cannot be determined as the graph lacks the lower asymptote, as shown in the lower graph. Conversely, the lowest concentrations should exert no effect compared to the no-compound control.
To learn more, visit
Sebaugh JL, Guidelines for accurate EC50/IC50 estimation
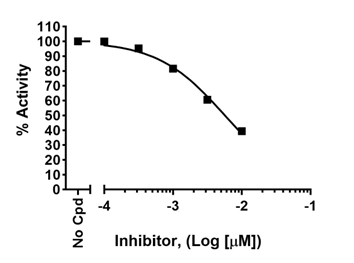
Background issues: how do I design my colorimetric assay when my compound has color?
If the compound being tested has color that interferes with the assay (for example, a yellow compound would interfere with colorimetric assays read at λ = 570–590 nm), a blank must be determined for each concentration of the compound and the dose-response signal must be normalized against the blank for each compound concentration.
Why is the apparent IC50 value in the validation experiment different from the published value for this compound?
First, let’s define what “different” means for IC50 values. A 2 or 3-fold variation is usually considered within range and The IC50 would not be considered different.
Next, remember that IC50 values are not constants. They differ depending on the type of assay and the parameters of the assay. Assay sensitivity is one of the critical parameters affecting the measured IC50. Thus, IC50 values obtained using a colorimetric assay kit will not be similar to IC50 values obtained using Mass Spec. In addition, within the same assay format, enzyme to substrate ratio, time of reaction, and whether the compound is pre-incubated or not with the enzyme prior to reaction initiation, among other parameters, may affect the value obtained.
When comparing compound efficacy in drug discovery campaigns, it is critical to use the same assay format and the same experimental conditions for all compared compounds. Ideally, the enzyme used throughout the campaign should be standardized for specific activity.
What is a Fluorescence Polarization (FP) assay?
Fluorescence Polarization (FP) is based on measuring changes in light polarization emitted by a fluorescent probe in a sample. It is quite different from fluorescence intensity, which measures the intensity of emitted light at a specific wavelength. FP is widely used to monitor molecular interactions in solution. FP is a complex technique that requires careful design, involves somewhat complicated calculations, and uses a specific instrument. It is, therefore, important to understand the principles underlying FP technology and FP-based experiments. An overview of the technology, principles of FP-based experiments, advantages and considerations, can be found here [
FP principles].
What type of instrument is needed?
A “standard” fluorescence plate reader measuring fluorescence intensity at a given wavelength will not capture FP data. The assay requires the use of an instrument capable of excitation with polarized light and capable of measuring fluorescence intensity on two different planes. For optimal experimental design, it is important to read the protocol provided with the kit all the way through before starting the experiment.
Why kind of assay plate is needed to perform an FP assay?
FP assays are performed in a black low-binding assay plate. Our kits include one assay plate per kit. The plate catalog number is indicated in the datasheet, should you need to purchase additional plates.
How sensitive is an FP assay?
FP technology measures fluorescence intensity emitted by the fluorophore in the two planes of light that are parallel and perpendicular relative to the plane of excitation. The degree of fluorescence polarization is defined as (P). Most instruments display fluorescence polarization in units of mP in which 1 mP = 1000 P.
Theoretical P values range from –0.33 to 0.5 (–330 to 500 mP), while experimental data typically range from 10 mP to around 300 mP.
FP instruments can achieve very precise measurements (± 2 mP).
Are the FP assay kits suitable for high throughput screening?
FP assay kits are ideal for high throughput screening because they are designed for small volumes and no wash steps are necessary. Most of our kits exist in a 96 well format, although some have also been optimized for use in a 384 well format.
What controls are needed to run a successful FP assay?
Several controls are needed to perform a successful FP assay. Controls are described in the protocol provided with BPS Bioscience assay kits.
-
Blank: the blank is meant to measure the background signal from the buffers used in the reaction. If studying a compound dissolved in an organic solvent, the same concentration of solvent should be present in all the controls. The Blank value is subtracted from all other values.
-
Low FP: internal control, calibration for the lowest signal allowed by the particular assay being performed. For example, a condition in which all the fluorescent tracer is in free form.
-
High FP: internal control, calibration for the highest signal allowed by the particular assay being performed. For example, a condition in which all the fluorescent tracer is in bound form.
-
Experimental control: a condition lacking the compound under study. If the compound is an agonist, the “no compound” condition is the negative control. If the compound is an inhibitor, the “no compound” condition is the positive control, and this value should be set as 100% for calculations.
For enzymatic reactions that require an agonist to work, keep in mind that the agonist should be added in all conditions except the negative control.
Are FP assay kits suitable to use with crude biological matrices?
FP assays may allow the interrogation of complex solutions (for example a urine sample) as long as they are well defined and free of contaminants. However, the purity and quality of the samples are critical. Potential interference in light scattering can be caused by many large molecules such as cell or membrane debris. For this reason, crude cell lysates, cell culture supernatants and other rudimentary extracts should not be used in this type of assay. The presence of contaminants with high background fluorescence or with non-specific trapping ability is likely to result in high noise-to-signal ratios or to otherwise interfere with the signal.
The specificity of our kits is determined by the purified enzyme. If the sample contains any trace of another enzyme that can modify the substrate, the kit will not function properly as the trace enzymatic activity will interfere.
Can I add BSA to my sample?
Bovine Serum Albumin (BSA) may interfere with some fluorescence kits. If BSA is needed, it is important to evaluate its effect by comparing polarization of the fluorescent tracer with or without BSA in the assay optimization phase.
A few assays developed by BPS bioscience contain BSA and have been optimized accordingly.
What can cause high background fluorescence signals?
An unacceptably high background signal (provided by the “Blank”) can be caused by contaminants in the buffers. Attention to raw materials, cleanliness of mixing and storage vessels, and buffer preparation methods should reduce high background signals. High backgrounds can also be caused by solvents* that fluoresce at the wavelength of interest, which would be indicated by an unusually high signal in the “blank”. When using an uncommon diluent for the first time, it can be useful to compare the signal with and without diluent to assess any effect of the diluent on the background signal.
*Solvents: used to dissolve a compound that is not soluble in aqueous buffers. Typical organic solvents include DMSO, ethanol or dimethylfluoride.
What are contraindications?
Contraindications are substances that interfere with the assay reaction. Anytime BPS Bioscience’s scientists observe that a solvent interferes with an assay, such as DMSO at a concentration of >1%, the observation will be indicated in the datasheet. Look for the “contraindications” section that will indicate known buffer or solvent interferences. Please keep in mind that BPS Bioscience does not test all solvents.
Some of the enzymatic assays can be impaired by specific buffers (for example phosphate-containing buffers interfere with Phosphodiesterase kits).
What is the “Gain” value setting?
The "Gain settings" are instrument-specific. Related information is usually found in the instrument owner’s manual. Most instruments have software programs that automatically adjust the gain settings based on inputs provided by the user when selecting the FP mode.
Typical settings:
-
Use the Blank subtraction setting. Select Blank reduction option for calculation of mP values. As a result, no value should appear in the Blank samples since the Blank is subtracted from all values obtained from the assay plate.
-
Input the G factor. The G factor is instrument specific and can vary from instrument to instrument. The G factor is typically between 0.5-1.5, may be provided by the owner’s manual or may be automatically computed by the instrument. If manual input is required by the instrument, determine the G factor as detailed below.
-
Select the low reference control. This will be the background signal when calculating the signal to background ratio.
-
No negative mP values should be obtained. Negative values are obtained when the perpendicular fluorescence readings are greater than the parallel fluorescence readings, which is caused by incorrect settings.
What is the G factor?
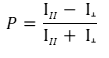
The P equation assumes that light is transmitted equally well through both parallel and perpendicular channels. In practice, this is generally not true and a correction must be made. This correction factor is called the "G Factor"; it is specific to the instrument and has to be determined by the user. Modern instruments usually have the G factor pre-calculated, so refer to your instrument instructions to determine how to set or calculate the G factor.
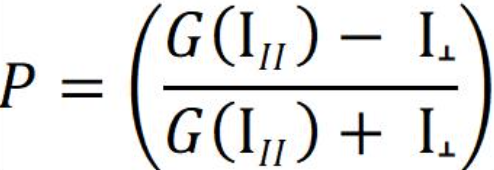
If you do need to calculate the G factor:
Use the theoretical mP of your fluorophore in the equation above (for example, the theoretical mP = 27 or P = 0.027 for fluorescein and Texas Red), together with the measurements for both the parallel and the perpendicular channels obtained from the free fluorophore at a concentration that gives counts well above the background “blank”. The background needs to be subtracted from both the parallel and the perpendicular fluorophore-only measurements. From the equation above, calculate:
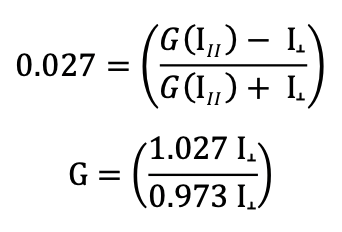
How do I determine the G factor when measuring FP?
The user may ignore the G-factor when all experiments are performed using the same instrument, since the G-factor is instrument-dependent.
Where needed, the G-factor should be set before measurements are performed. Some manuals will clearly say what the G-factor is. For example, our scientists use a Tecan fluorescent plate reader which has a G value set to 22 mP.
However, the G-factor may need to be figured out by the investigator when not clearly indicated by the manufacturer. The instrument manual will contain information about how to establish the G-factor.
How do I calculate Fluorescence Polarization results?
Instruments give measurements in milli-Polarization = mP.
Calculate ΔmP for all samples:
ΔmP = (mP value of the sample) – (mP value of the Reference control)
Where mP refers to milli-Polarization values provided by the instrument and Reference control is the mP value obtained in the condition containing only the fluorescent probe (a condition in which the probe is in free state).
What if the Alpha-counts signal of positive control reaction is same as “Blank” value?
Possible Causes and Solutions
Enzyme has lost activity: Enzyme loses activity upon re-peated freeze/thaw cycles. Use fresh JMJD2A, BPS Bioscience #50123. Store enzyme in single use aliquots. Increase time of enzyme incubation. Increase enzyme concentration.
Streptavidin Donor beads or anti-mIgG acceptor beads fail to show significant signal: Reorder Streptavidin Donor beads or anti-mIgG acceptor beads from Perkin Elmer.
Incorrect settings on instruments: Refer to instrument instructions for correct settings to increase sensitivity of light detection.
The Alpha-counts signal is erratic or varies widely among wells. What could be wrong?
Inaccurate pipetting/technique: Run duplicates of all reactions. Use a multichannel pipettor. Use master mixes to minimize errors.
What is a TR-FRET assay?
Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) is commonly used to analyze the binding of two interacting molecules. The technology is a combination of Time-Resolved Fluorescence and Förster’s Resonance Energy Transfer (FRET), a phenomenon in which a light-excited fluorophore can transfer its absorbed energy to a nearby acceptor fluorophore. An eBook describing the principle of TR-FRET and examples is available to download or print [
eBooks].
What are the advantages of TR-FRET experiments?
TR-FRET is an ultra-low background technique allowing the measure of any reaction in which two labeled entities come in proximity. The main drawbacks of the technique are that it requires two optimized labeled entities, in addition to exhibiting a low dynamic range. However, these drawbacks are offset by several advantages:
-
Simple protocols, fast assays
-
Small volumes
-
Homogeneous: no need for washing steps or for physical separation from the unbound entities
-
Robust, sensitive signal
-
Ultra-low background with high signal to noise ratio
-
Stable signal: use of lanthanide donor fluorophores minimizes photobleaching
The growing commercial availability of ready-to-use TR-FRET immunoassay kits has opened the technique to mainstream use. BPS Bioscience offers >100 assay kits for drug discovery in the TR-FRET format with new products developed regularly. Optimized, validated, high quality assay kits ensure reliable results, quickly.
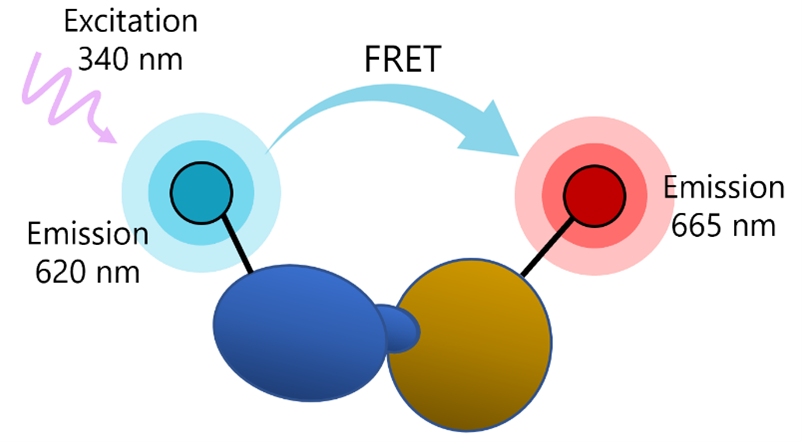
What is Förster’s Resonance Energy Transfer (FRET)?
FRET is a phenomenon in which two fluorophores emitting at different wavelengths are coupled: the donor fluorophore excited by a high energy source transfers energy (not light) to an acceptor fluorophore. This results in excitation of the acceptor and fluorescence emission at another wavelength, which can be detected by a fluorescence reader.
The transfer of energy between the donor and the acceptor depends on physical proximity (<10 nm) and decreases rapidly with distance. Thus, partner molecules distributed in a solution are sufficiently far apart and FRET does not occur. Upon interaction, the partners complete the FRET pairing as they are now in proximity to each other.
What does Time-Resolved Fluorescence (TRF) mean?
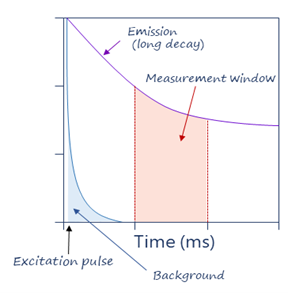
Classic fluorescence intensity uses short-lived fluorophores such as fluorescein, with an emission speed in the order of the nanoseconds. Excitation and emission occur at specific wavelengths that can be differentiated by a fluorescence reader. However, excitation and emission happen at the same time. If there is any amount of spectral overlap between excitation and emission, as there usually is, the reader will capture some of the excitation fluorescence, resulting in background signal and low signal-to-noise ratios.
TRF solves this by using long-lived inorganic fluorophores as donors and adding a time delay between excitation and measurement, which means that the excitation signal is gone by the time of the measurement, which decreases background signals. TRF also uses excitation pulses (not continuous excitation), so that a series of measurements are repeated over time. TRF also eliminates transient background fluorescence generated from sample components such as buffers, proteins, and chemicals, which hinders classic FRET methods. Thus, TRF technology accounts for the ultra-low background advantage of TR-FRET.
What are the most common fluorophores?
Ideal fluorophores have high signal intensity, are highly stable, and offer excellent signal-to-noise ratios. Fluorophores commonly used are “Lanthanide probes” which are metal ions referring to elements Cerium to Lutetium in the periodic table. The most used are Europium and Terbium, which fluoresce over milliseconds instead of nanoseconds.
What type of instrument is needed?
A fluorescent plate reader capable of measuring Time Resolved-Fluorescence Resonance Energy Transfer is needed for these experiments.
How is measurement made?
In practice, a comparison measurement of the two emitted wavelengths over time is calculated for a TR-FRET response.
Therefore, two sequential measurements are conducted. For example, Tb-donor emission should be measured at 620 nm followed by dye-acceptor emission at 665 nm. Data analysis is performed using the TR-FRET ratio (665 nm emission/620 nm emission).
To calculate the percentage activity, subtract the Blank value from all other FRET values. It is expected that Blank and Negative Control have a similar value. The FRET value from the Positive Control can be set as one hundred percent activity as it corresponds to the maximum activity.
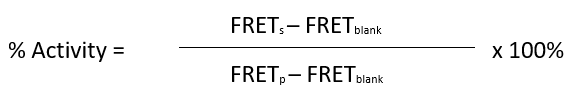
Where FRETs = Sample FRET, FRETblank = Blank FRET, and FRETP = Positive control FRET.
Why do I obtain a low luminescence signal in my positive control compared to my test inhibitor when using Kinase-Glo® MAX reagent in your kinase assay kits?
Kinases use ATP to add a phosphate group onto a substrate protein. The phosphorylation reaction, therefore, depletes ATP and generates ADP.
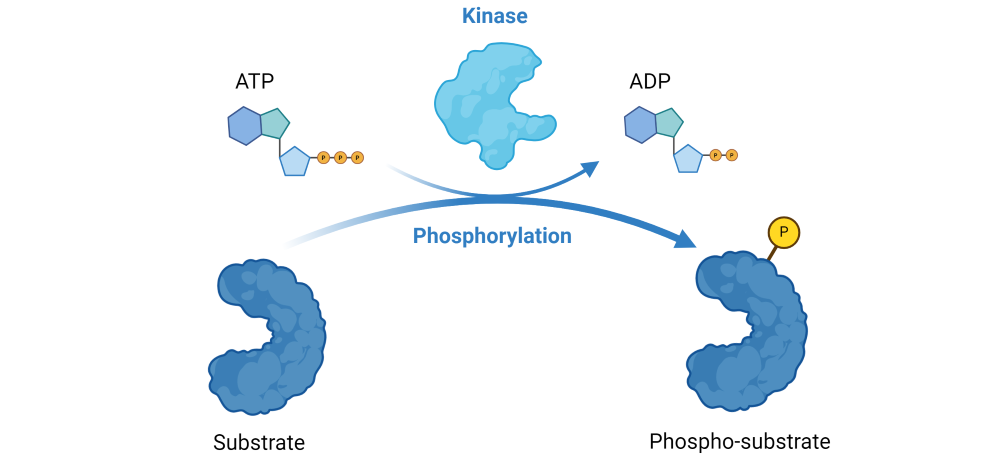 Figure 1: Illustration of the phosphorylation reaction catalyzed by kinases.
Figure 1: Illustration of the phosphorylation reaction catalyzed by kinases.
The Kinase-Glo® MAX reagent is added as a detection reagent that quantitatively measures the remaining, unused ATP. The reagent is linear up to 500 µM ATP.
Since the luminescent signal correlates with the amount of remaining ATP, it is inversely proportional to kinase activity. Thus, a decrease in the luminescent signal corresponds to higher kinase activity, while an increase in the signal corresponds to lower kinase activity.
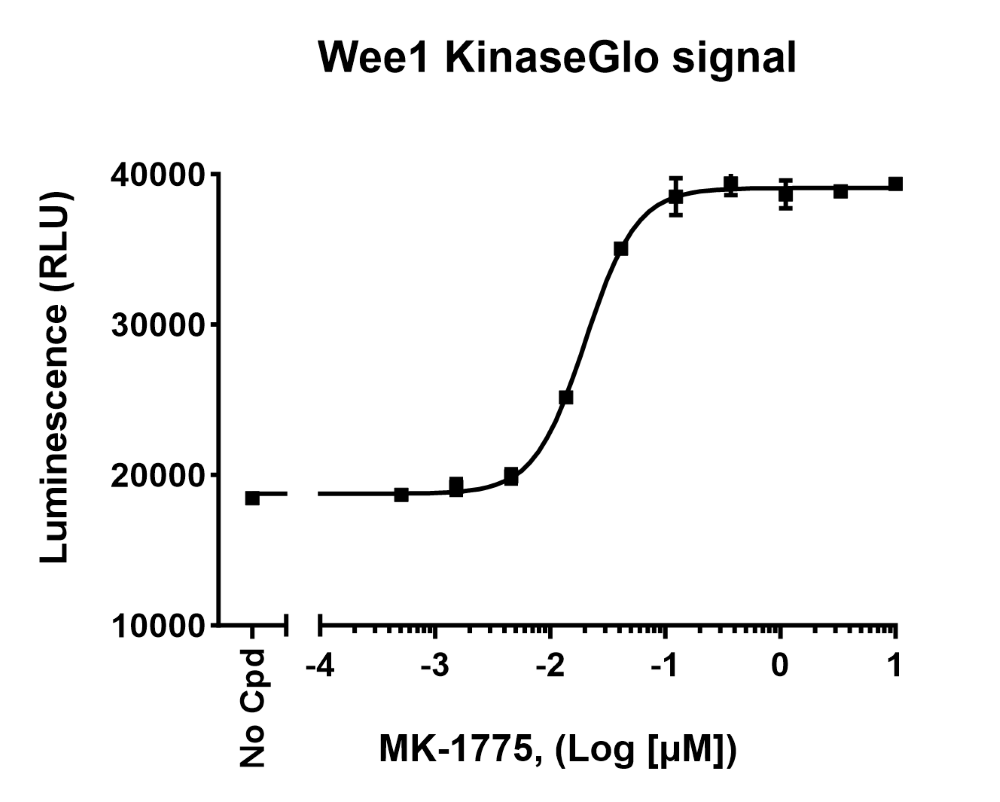 Figure 2: Raw luminescence data obtained by measuring Wee1 kinase activity in the presence of increasing concentrations of inhibitor MK-155, using Kinase-Glo® MAX reagent (Wee1 Kinase Assay Kit (#79909)).
Figure 2: Raw luminescence data obtained by measuring Wee1 kinase activity in the presence of increasing concentrations of inhibitor MK-155, using Kinase-Glo® MAX reagent (Wee1 Kinase Assay Kit (#79909)).
Validation graphs shown in the assay kit datasheets show results expressed as percent of kinase activity, in which kinase activity in the absence of inhibitor (“positive control”) is set to 100%.
Results are calculated by averaging replicates and subtracting the blank from all experimental values. Thus, for each “test inhibitor” concentration, calculate:
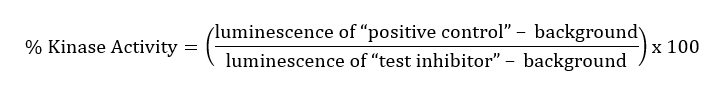
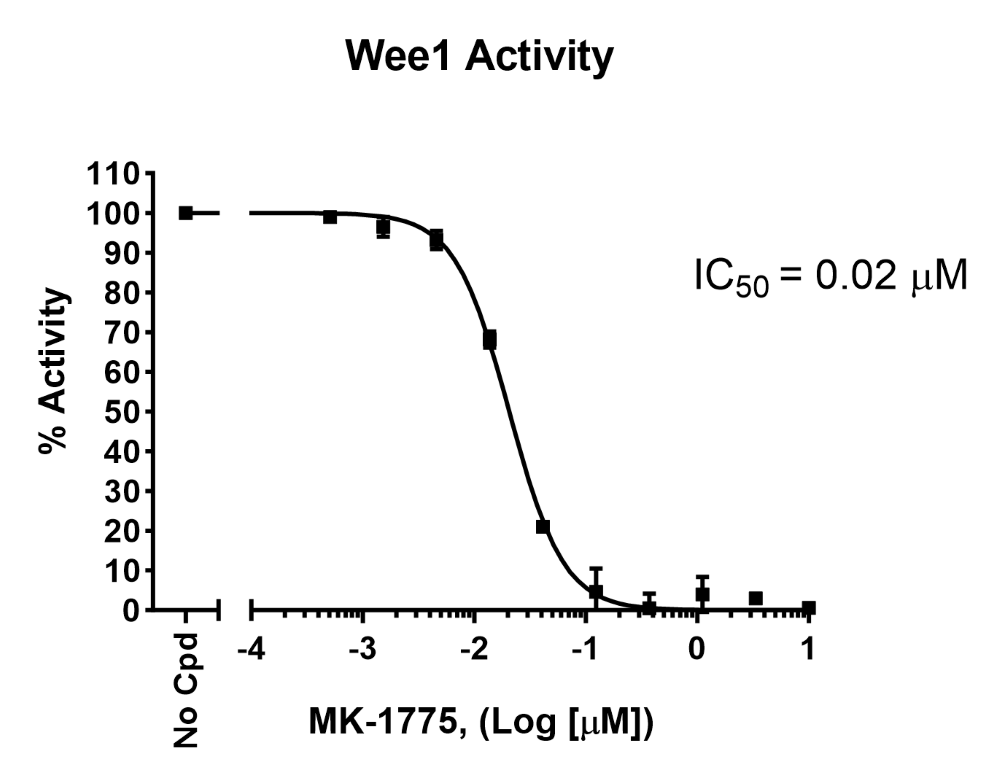 Figure 3: Inhibition of Wee1 kinase activity by MK-1775 measured using Wee1 Kinase Assay Kit (#79909). Results are expressed as percent of activity (positive control without MK-1775 set to 100%).
Figure 3: Inhibition of Wee1 kinase activity by MK-1775 measured using Wee1 Kinase Assay Kit (#79909). Results are expressed as percent of activity (positive control without MK-1775 set to 100%).
How should I store the viral particles (lentivirus, AAV or VSV) that I just received?
Our viral products are shipped with dry ice. If there is no dry ice left in your shipment upon arrival, please let us know at once. For long term storage, it is recommended to store the viral particles at -80°C. Avoid repeated freeze-thaw cycles as titers can drop significantly with each freeze-thaw cycle.
I ordered off-the-shelf viral particles and kept them at -80°C as instructed. How do I thaw the virus?
Thaw at room temperature and add directly to the cells. If an intermediate dilution is needed, the virus can be diluted in the cell culture medium at room temperature.
Titers can drop significantly with each freeze-thaw cycle. If the virus preparation is going to be used multiple times, it is best to prepare very small aliquots and freeze them immediately. Avoid multiple freeze/thaw cycles.
What are the advantages of lentivirus transduction over lipid-mediated transfection?
Lentivirus transduction is a powerful tool for introducing genes into cells and has a few advantages over lipid-mediated transfection, particularly in terms of efficiency, stability, cell type versatility, and cargo capacity.
- Efficiency: Lentivirus transduction is more efficient than lipid-mediated transfection, meaning that it can introduce genetic material into a higher percentage of cells. This is because lentiviruses have evolved to efficiently enter and integrate into the host cell's genome. Thus, they are especially useful to transduce cells that are somewhat resistant to lipid-mediated transfection.
- Stable integration: Lentiviruses can integrate into the genome, resulting in stable and long-lasting expression of the introduced gene. Lipid-mediated transfection typically results in transient expression and integration is a rare event (even when using linearized DNA).
- Versatility: Lentivirus transduction is used to introduce genes into a wide range of cell types, including both dividing and non-dividing cells. In contrast, lipid-mediated transfection is less efficient in primary cells or non-dividing cells.
- Lower toxicity: Lentivirus transduction is less toxic to cells than lipid-mediated transfection.
- Larger cargo capacity: Lentiviruses can carry larger genetic payloads than lipid-mediated transfection methods, allowing for the introduction of more complex genetic constructs.
What is a pseudovirus?
The most commonly used lentivirus is based on the HIV virus that infects human lymphocytes through the specific interaction of its envelope protein gp120 with the CD4 receptor. In biotechnology, gp120 is replaced by another envelope glycoprotein, typically VSV-G (Vesicular stomatitis virus G protein), which binds to the human LDL receptor (low-density lipoprotein receptor) expressed on most cell types. An HIV-based lentivirus containing an envelope protein other than gp120 is called a pseudotyped virus, or pseudovirus. Pseudoviruses in which the envelope protein has been replaced by the SARS-CoV-2 Spike protein will bind to human ACE2 and are useful to study Spike/ACE2-mediated infection.
View Products
What are bald lentiviruses?
Bald lentiviruses are pseudovirus particles in which the envelope protein has been removed entirely. Therefore, the bald pseudovirus cannot interact with host cells in a receptor-specific fashion. Bald lentiviruses serve as a negative control (or blank) in infection experiments.
View Products
What are reporter lentiviruses?
Reporter lentiviruses contain a plasmid to express a reporter gene such as GFP or Luciferase in the target cell. This reporter gene can be under the control of a constitutive promoter (e.g. CMV) or under the control of a promoter containing a specific response element to study the activation of a particular transcription factor or signaling pathway. The reporter gene is transduced when the lentivirus enters the target cell and the resulting expression of the reporter protein can be detected by fluorescence (GFP) or luminescence (luciferase activity). SARS-CoV-2 Spike reporter pseudoviruses can be used to quantify Spike/ACE2-mediated infection.
View Products
What safety level do you need to use BPS lentiviruses?
A biosafety level 2 facility is sufficient to use BPS lentiviruses. Our replication-incompetent lentivirus particles, produced from packaging cells, contain minimal genetic information that includes the gene of interest but does not include
viral genes involved in packaging. Therefore the virus packaging genes are not expressed in the transduced cells and cells infected with our viruses cannot produce new viral particles.
What types of cells can you infect with a lentivirus?
Lentiviruses infect most cell types and can transduce dividing cells as well as quiescent cells, whether they are immortalized or primary cells, which makes them particularly suitable to use with cells that are difficult to transfect using other methods.
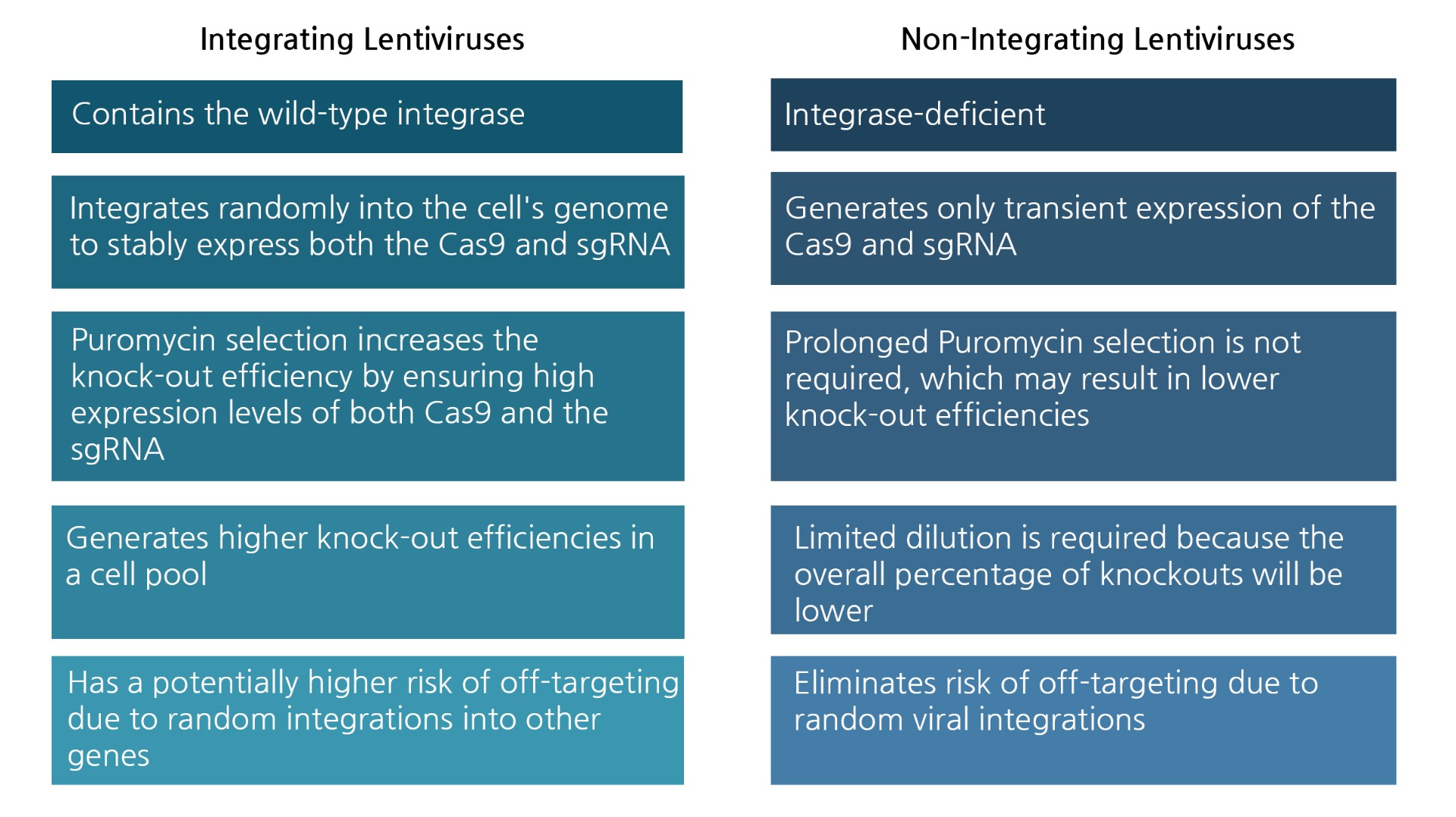
What is a non-integrating lentivirus?
A lentiviral genome integrates in the host DNA using an enzyme called retroviral integrase. This enzyme has been deactivated in BPS Bioscience’s integration-deficient lentiviruses, therefore the DNA carried by the virus cannot integrate into the host DNA. The lack of integration prevents possible off-target effects resulting from the random genetic disruption in CRISPR/Cas9 knock-out cells. They are used for transient transduction or as control for integrating viruses.
 View Products
View Products
What is an MOI?
Multiplicity Of Infection (MOI) is the ratio of the number of infective virions to use per number of target cells. For example, if 100,000 infective particles are added to 1 million cells, the MOI is 0.1. The optimal MOI needs to be determined for each cell line. Optimization is easily carried out using a GFP reporter lentivirus (BPS Bioscience, #79979). Suggested MOIs for commonly used cells are indicated in the following table:
| Cell Line |
HEK293 |
HeLa |
HCT116 |
Jurkat |
MCF7 |
MDA-MB-231 |
A549 |
| Suggested MOI |
5 |
3 |
5 |
10 |
2 |
1 |
5 |
What are good cellular models to use BPS Bioscience Spike (SARS-CoV-2) pseudoviruses?
Spike interacts with human cells by binding to membrane receptor ACE2 (angiotensin converting enzyme 2), therefore cells expressing high levels of ACE2 are good models. For best results we recommend using ACE2-overexpressing cells such as ACE2-HeLa, ACE2-CHO and ACE2-HEK293 cells, in which ACE2 overexpression has been validated by Flow Cytometry. Vero E6 cells are also commonly used for coronavirus studies and do express ACE2 endogenously. However, based on experiments performed by BPS scientists, ACE2-HEK293 cells provide greater transduction when compared to Vero E6 cells. To optimize Vero E6-based experiments, BPS recommends using TMPRSS2-Vero E6 cells. These cells express serine protease TMPRRS2 that cleaves the S2 subunit of Spike and promotes membrane fusion and viral entry into the host cell.
View Products
What is the best AAV serotype for my cell type?
So far, 11 AAV serotypes have been isolated from humans and non-human primates. Each serotype shows preferential binding for particular cell types and tissues, as determined by the binding affinity of the capsid proteins to receptors on the cell surface (see tables below). Scientists can use this tropism to efficiently target specific cell types.
Genetically engineered AAV serotypes have been developed to further increase tissue tropism and transduction efficiency. For example, AAV-DJ is a synthetic serotype made from eight different wild-type AAV serotypes (AAV2, 4, 5, 8, 9, avian, bovine, and goat AAV) using DNA shuffling. The AAV-DJ serotype can infect a broad range of cell types and has better transduction efficiency in vitro and in vivo compared to wild-type serotypes.
A survey of in vitro transduction efficiency for serotypes 1 to 9 has been published (
Ellis BL, et al. 2013)
|
Serotype
|
Tissue
|
|
AAV1
|
CNS, Heart, and Skeletal
Muscle
|
|
AAV2
|
CNS and Kidney
|
|
AAV3
|
Liver
|
|
AAV4
|
CNS and Lung
|
|
AAV5
|
CNS and Lung
|
|
AAV6
|
Lung and Skeletal Muscle
|
|
AAV7
|
Liver and Skeletal
Muscle
|
|
AAV8
|
CNS, Heart, Liver, Pancreas, and Skeletal
Muscle
|
|
AAV9
|
CNS, Heart, Liver,
Lung, and Skeletal Muscle
|
What safety level do you need to use AAVs?
AAVs require the use of a Biosafety Level 1 facility. BPS Bioscience recommends following all local, federal, state, and institutional regulations and using all appropriate safety precautions.
AAVs are replication-defective, non-pathogenic, and have not been associated with any human disease. Wild-type AAVs integrate into the host genome at negligible frequencies. When it occurs, integration is predominantly at the “safe harbor” site AAVS1 on the human chromosome 19. However, recombinant AAV vectors exist as episomes inside the cells and therefore do not disrupt the genome.
What is the titer of the supplied AAVs?
Our AAVs are supplied as two vials (50 µl x 2) of AAV at a titer ≥1 x 1012 vector genomes (vg)/ml. Therefore, each vial contains approximately 50 billion vector genomes. The titer is determined by qPCR and varies with each lot; the exact value is provided with each shipment.
What kind of quality control do you perform on AAVs?
Each AAV preparation is purified by iodixanol density gradient and ultra-centrifugation. The titer is determined by SYBR Green qPCR. Titers vary with each lot and the exact titer value is provided with each shipment. The purity of the AAV particles is determined for each lot by staining on a 4-20% SDS-PAGE gel using One-Step Lumitein™ UV Protein Gel Stain (Biotium #21005) and is guaranteed to be greater than 90%. If the AAV contains a fluorescent reporter gene, it is functionally confirmed by transducing HEK293 cells and detecting the resulting fluorescence using a fluorescence microscope. For luciferase reporter AAVs, HEK293 cells are transduced, and the luciferase activity is measured using the ONE-Step™ Luciferase Assay System (BPS Bioscience #60690). Finally, SaCas9 AAVs are tested by transducing HEK293 cells and confirming HA-tagged SaCas9 expression by Western blot using an anti-HA-tag antibody.
How do I thaw my AAVs?
Keep AAV frozen -80°C until use. Thaw the AAV particles on ice. Quickly centrifuge the tube to recover its full content. Gentle vortex mixing of the tube is also acceptable. The viral particles may be diluted in cell culture medium if desired or can be added directly to the cells. Alternatively, AAVs can be diluted in Phosphate Buffered Saline (PBS) for in vivo use.
AAVs are sensitive to freeze/thaw cycles. The viral titer will drop sharply with each freeze/thaw cycle.
Can I use AAVs with confluent cells?
Most adherent cells are more efficiently transduced when they reach approximately 70% confluency. However, AAVs can also be used with cells at >70% confluency provided that the experimental conditions have been optimized accordingly (for example, by increasing the Multiplicity of Infection).
Can I use AAVs with Primary cells?
Yes. In fact, AAV transduction works better than other methods of transfection or viral transduction when using primary and non-dividing cells, including induced pluripotent stem (iPS) cells and iPS-derived cells.
Can I generate a stable cell line using AAV?
No. AAVs do not integrate into the genome and are not suitable to generate stable cell lines. Transduced genes are maintained in the cells as episomes and can be expressed for up to 6 months or longer in non-dividing cells, or up to 3 months in dividing cells (depending on the cell type and MOI used). However, expression is diluted over time in dividing cells.
What is T2A?
The T2A linker is a self-cleaving peptide derived from Thosea asigna virus 2A that is used to express two reporter proteins at equal levels of expression. For example, our dual-reporter AAV systems such as AAV luciferase-eGFP use the DNA construct “Luciferase-T2A-eGFP” under the control of a single promoter. The product of this DNA construct is a unique transcript which encodes the protein “Luciferase-T2A-GFP”. Linker T2A undergoes self-cleavage, leading to the release of luciferase and eGFP as two separate proteins in roughly equal proportions.
Can I use AAVs from BPS Bioscience for in vivo experimentation?
Yes, BPS Bioscience’s AAVs can be used in animal research and may be injected into laboratory animals. Our purification process removes empty capsids from AAV preparations and guaranties high quality AAV particles. If necessary, users may request further purification or testing as a
custom order.
Please note that our products are not for use in humans or for diagnostic or therapeutic applications.
How do I obtain the best possible efficacy of transduction?
Reporter AAVs can be used to optimize transduction efficiencies in your cell line of interest. Particle-to-cell ratio is the most critical parameter to vary when optimizing protein expression in the target cells. Adjusting the incubation volume is the next best parameter to optimize. Adjusting the temperature or the cell density (if adherent cells are being transduced) may also improve transduction efficiencies. For adherent cells, we recommend a confluency of about 70%.
What are the SaCas9 AAVs?
SaCas9 (Staphylococcus aureus CRISPR-associated protein 9) has demonstrated high nuclease efficiency in mammalian cells, and its small size makes it ideal for packaging into AAV. SaCas9 recognizes a longer protospacer adjacent motif (PAM) site, 5’-NNGRRT-3’, than the more traditional SpCas9 (Streptococcus pyogenes CRISPR-associated protein 9). These AAV particles constitutively express SaCas9 under the control of a CMV promoter. They are used to constitutively express SaCas9 in the target cells to genetically engineer the cells using CRISPR/Cas9 technology.
What information is needed when ordering a custom AAV design or custom AAV packaging service?
Our team of scientific experts is available to help with the
custom design or your AAVs. To expedite the process, these are the points to consider.
For custom AAV design:
- If you have a gene template, please provide your gene template to us.
- If you would like us to synthesize a gene without a template, please provide the gene number, sequence map, species, and gene region.
- To generate AAVs for the manipulation of gene expression using techniques such as RNAi or CRISPR, please provide us with target gene information such as database accession number, species, and gene length (ideally, under 3.2 kb).
- Please indicate if you prefer a specific promoter to be used in the construct.
- Please indicate if you would prefer to use a particular reporter gene.
- If two or more genes will be co-expressed, please provide all relevant information for each gene.
For custom packaging services:
- Please indicate which AAV serotype you will use. Our team is available to guide your selection if needed.
- If it is unknow which serotype is capable of infecting your cells, we offer reporter AAVs for the comparison of various serotypes to help you determine the optimal serotype for your experiments.
- If providing your own plasmid for packaging, confirm ITR spacing and fully sequence the plasmid to avoid possible complications associated with common mutations driven by the presence of the ITRs. Please send us the vector map and the full sequence of the plasmid.
- When placing your custom order, please indicate the desired amount, titer, and packaging requirements for your deliverable AAV.
What kind of services does BPS Bioscience offer?
Development services: we develop custom recombinant proteins and new assays, generate cell lines overexpressing a target of interest or containing a reporter gene, and develop cells containing the CRISPR-mediated genetic modification(s) of your choice. In addition, we can assist you with the design and validation of a tailored chimeric antigen receptor (CAR) of interest, or with AAV (Adeno Associated Virus) or lentivirus development. Visit
BPS Bioscience Services for more information.
Screening and Profiling:
- Biochemical Screening and profiling services: our full portfolio of >600 assays is available as a service: we perform screening, IC50 determination, and comparisons of drug efficacy across protein families. Visit Screening and Profiling Services to learn more.
- Cell-based screening services: characterize or titrate your drug in one or several cell lines. Cellular studies provide a biological context, allowing for more informed decisions about which compounds to advance to development. Visit Immunotherapy Screening Services
Can I receive guidance on development projects?
Absolutely. Our team of scientists will communicate with you throughout the project's design to ensure that the results correspond to what you need.
Does BPS Bioscience use a reference compound as internal control?
Yes. Our assays are validated with an internal control using a commercially available inhibitor whenever available. When several compounds have been used in an assay (visit the assay page), you can choose between them. Some assays may not have a commercially available reference inhibitor.
Alternatively, you may request that we use a reference compound of your choice. Please send your reference compound to us together with your test compounds, indicating clearly which compound is your reference.
How do I submit a request?
Please use the
contact form at the bottom of the Contact Us page, where you can provide your contact information and details about your project.
Make sure to include which target protein(s) you wish to study, how many compounds you have to test, and whether you need a single concentration assay or a full IC
50.
Remember that if several reference compounds are available for an assay, you need to indicate which one(s) should be used as internal control(s). If you wish to use a different reference compound than the compound used by BPS, please send it to us together with your test compounds, indicating clearly which compound is your reference.
Alternatively:
- Contact your Sales representative (US) at [email protected]
- International customers (non-US) should contact their local distributor. Contact your distributor (international). For a list of International Distributors, visit Worldwide Distributors
How should I prepare compounds?
Prior to shipping your compounds, please review our
Compound Submission guidelines. Fill out a “Compound Submission Form” and include it with your compounds when shipping,
in addition to sending it to the email address provided on the form.
Biochemical Assay
Compound Submission Form
Cellular Assay
Compound Submission Form
Remember that if several reference compounds are available for an assay, you need to indicate which one(s) should be used as internal control(s). If you wish to use a different reference compound than the compound used by BPS, please send it to us together with your test compounds, indicating clearly which compound is your reference.
Compound Preparation:
- Biochemical assays: Prepare at least 50 µl of compound solution, at a concentration that is 100-fold higher than the highest concentration to be tested if the diluent is DMSO (dimethyl sulfoxide).
- Cellular assays: Prepare at least 50 µl of compound solution, at a concentration that is 1000-fold higher than the desired final concentration if the diluent is DMSO.
As a rule, compounds in solution are preferred over powder form. We do accept dry compounds as long as their molecular weights are clearly indicated, to ensure accurate solutions are prepared. If sending your compounds in powder form, please send 1 to 3 mg, depending on the molecular weight of the compound. If we experience issues with compound solubility, unless informed otherwise, we will take the following steps to solubilize the compound:
- Further dilution (if possible)
- Sonication
- Heat at 50°C
BPS will store compounds for a maximum of 6 months after receipt.
Where do I send compounds?
Mail your compounds to:
BPS Bioscience
ATTN: Compound Submissions
6405 Mira Mesa Blvd., Suite 100
San Diego, CA 92121
Phone: 858-202-1401
Once the package has been shipped, notify us of the tracking number, so we can monitor the shipment and ensure your compounds arrive safely.
How are results communicated to me?
When a project has been completed, BPS Bioscience will provide you with an extensive report containing raw and analyzed data, graphs, and detailed protocols. Testing typically includes internal controls for inhibition. This report will be sent to you directly.
Can I have details on the protocol used for screening or profiling before ordering a service?
Yes. Our services correspond to existing assays, which can be viewed on our website. Open the datasheet corresponding to the assay you are interested in. All our assay kits are provided with detailed protocols and validation data, available from the datasheet.