Assay Kit Frequently Asked Questions
Do you sell inhibitors or agonists to use as internal controls?
Yes, we do sell support reagents such as inhibitors and agonists. Note that an internal control inhibitor or agonist may be included with some assay kits. Check in the table of “Assay Kit Components” of the product page.
What is the composition of your buffers and reagents?
In some cases, the composition of a reagent will be available from the web page and the technical datasheet corresponding to the product of interest. However, the composition of many of our reagents is proprietary. If you need to know whether a specific chemical is present in one of our proprietary reagents (for example, dithiothreitol), contact us with your question at [email protected].
What is a homogeneous assay?
An assay in which no wash steps are required to remove unbound molecules, since the presence of unbound molecules does not interfere with the reading. This reduces the number of steps necessary to perform the assay and makes it more amenable to high-throughput applications. Fluorescence Polarization, TR-FRET, AlphaScreen™, and Fluorogenic assays are typically homogeneous assays. ELISAs (Enzyme-linked immunosorbent assays) are heterogeneous assays with several wash steps required throughout.
Can I use cell lysates or other complex extracts with the kit?
No. Our biochemical assay kits are based on the validated activity of a recombinant, affinity-purified protein of interest. They are designed to directly test the effect of compounds on the target enzymatic activity, or on the binding between two partners. They are not designed to assay enzymatic activity present in a complex system such as cell lysates and we do not recommend it as it would not be possible to assess the specificity of the signal generated.
How do I store the kit components?
Store each kit component as indicated in the “Supplied Materials” table displayed in the datasheet or the “Format” table shown on the product page.
Most kits contain purified recombinant proteins that are very sensitive to temperature and to freeze/thaw cycles. Kits are shipped with dry ice to maintain the full activity of the kit components. Contact technical support at [email protected] if you notice an issue with the shipment.
Most kits contain purified recombinant proteins that are very sensitive to temperature and to freeze/thaw cycles. Kits are shipped with dry ice to maintain the full activity of the kit components. Contact technical support at [email protected] if you notice an issue with the shipment.
Where can I find the protocol of the assay kit?
Detailed assay protocols are provided in the datasheet corresponding to the assay kit. Datasheets are available on our website and can be accessed from the product page.
Always read the protocol carefully from start-to-finish before starting an experiment.
Always read the protocol carefully from start-to-finish before starting an experiment.
How do I know if BPS Bioscience has tested a reference compound in an assay?
The assay datasheets usually show all the results validated by our team of scientists and the protocols used to generate the data. Additional information about other researchers’ use of an assay may be found in publications citing our assay (look in the Citations section of the product page).
I spilled a reagent used in my assay kit. Can I order it separately?
Yes, some reagents are available separately. Contact your Sales Representative or send us a message providing the name and catalog number of the reagent at [email protected].
Do you sell inhibitors or agonists to use as an internal control in biochemical assays?
We often provide an internal control with our assay kit. In addition, we sell support reagents such as inhibitors and agonists.
Can I use the kit more than once?
If the assay plate is going to be used more than once, prepare enough reagents for this portion of the assay and aliquot the remaining undiluted reagents into single-use aliquots depending on how many times the assay plate will be used. Store the aliquots at -80°C or at -20°C as appropriate.
Keep in mind that some enzymatic activity will be lost with each freeze/thaw cycle.
Keep in mind that some enzymatic activity will be lost with each freeze/thaw cycle.
What precautions should I take using the proteins supplied with the kit?
The concentration of each protein is lot-specific and is indicated on the tube. Always verify the stock concentration before using the protein.
Thaw on ice and gently mix prior to use. DO NOT VORTEX. Perform a quick spin before opening to recover the full contents of the tube. Proteins should be kept on ice until use. If dilution is required, use the Assay Buffer provided with the kit. Discard unused diluted protein at the end of the experiment.
Thaw on ice and gently mix prior to use. DO NOT VORTEX. Perform a quick spin before opening to recover the full contents of the tube. Proteins should be kept on ice until use. If dilution is required, use the Assay Buffer provided with the kit. Discard unused diluted protein at the end of the experiment.
What is a stepwise dilution?
A stepwise dilution is used when the stock solution of a component, such as a protein, is provided at high concentration. It is especially recommended in instances when diluting the reagent in a single step would result in a very large volume and/or use most of the buffer.
For example, if the protocol requires diluting a reagent 1,000-fold in Assay Buffer, do NOT dilute 10 µl of reagent in 10 ml of assay buffer. Instead, dilute 10-fold by adding 10 µl of reagent to 90 µl of Assay Buffer, then dilute again 100-fold by adding 10 µl of the previous dilution to 990 µl of Assay Buffer.
For example, if the protocol requires diluting a reagent 1,000-fold in Assay Buffer, do NOT dilute 10 µl of reagent in 10 ml of assay buffer. Instead, dilute 10-fold by adding 10 µl of reagent to 90 µl of Assay Buffer, then dilute again 100-fold by adding 10 µl of the previous dilution to 990 µl of Assay Buffer.
How do I dissolve the test compound that I will use in the assay?
It depends on the compound. If it is water-soluble, dissolve in the Assay Buffer as recommended in the protocol and prepare serial dilutions in the Assay Buffer. Controls should use the Assay buffer as well.
If the compound is soluble in organic solvents, such as DMSO, dissolve in 100% DMSO (or other appropriate solvent) at a high concentration, then dilute in Assay Buffer. Keep in mind that if you are testing a serial dilution, the concentration of DMSO should be kept constant across the dilution and controls.
Note that most of our assays are validated using DMSO as an organic solvent. Validated experimental conditions are indicated in the datasheet. We recommend that you read the protocol carefully from start-to-finish before starting the experiment.
If the compound is soluble in organic solvents, such as DMSO, dissolve in 100% DMSO (or other appropriate solvent) at a high concentration, then dilute in Assay Buffer. Keep in mind that if you are testing a serial dilution, the concentration of DMSO should be kept constant across the dilution and controls.
Note that most of our assays are validated using DMSO as an organic solvent. Validated experimental conditions are indicated in the datasheet. We recommend that you read the protocol carefully from start-to-finish before starting the experiment.
Could some common chemicals affect my assay?
Some assays are sensitive to the presence of a particular chemical. Examples of chemicals or buffer components known to alter the performance of biochemical assays: DMSO >1%, strong acids or bases, ionic detergents, high concentrations of salt. When our scientists have determined that a chemical or experimental condition has unwanted consequences in an assay, it will be indicated on the datasheet.
What kind of plate should I use?
Assay kits include a 96-well or a 384-well plate to perform the experiment. Low-binding plates are used with homogeneous assays such as TR-FRET assays. White plates are used with luminescent or chemiluminescent assays, whereas black plates are used with fluorogenic assays.
Some ELISA plates may be pre-coated, such as nickel-coated plates to capture His-tagged proteins or Avidin-coated plates to capture biotinylated proteins (NeutrAvidin™ Plate #79512). Be sure to store the plates as directed in the datasheet.
Some ELISA plates may be pre-coated, such as nickel-coated plates to capture His-tagged proteins or Avidin-coated plates to capture biotinylated proteins (NeutrAvidin™ Plate #79512). Be sure to store the plates as directed in the datasheet.
What is an IC50?
IC50 (Inhibitor Concentration 50) is a quantitative measure of the potency of a compound in inhibiting a biological or enzymatic function, in vitro. Specifically, it measures the concentration at which the compound inhibits a given biological or enzymatic process by 50%.
What is the difference between IC50 and EC50, and when do I use one or the other?
IC50 and EC50 are used in dose-response curves to determine the potency or effectiveness of a substance. The lower the value the greater the potency as it means that lower concentrations of the substance achieve the 50% effect.
IC50 (Inhibitory Concentration 50) represents the concentration of compound required to inhibit an effect by 50%. Therefore, it is used in studies involving inhibitors or neutralizing peptides and antibodies. For instance, in drug development, IC50 values are used to assess the potency of inhibitors against enzymes or to neutralize ligand/receptor binding.
EC50 (Effective Concentration 50) represents the concentration of compound required to produce 50% of a desired effect (e.g., 50% of the maximum achievable response). It can be used to measure the potency of agonists such as a receptor ligand, hormone, or neurotransmitter. It is also used in pharmacology studies to determine the potency of a drug in eliciting a therapeutic effect.
IC50 (Inhibitory Concentration 50) represents the concentration of compound required to inhibit an effect by 50%. Therefore, it is used in studies involving inhibitors or neutralizing peptides and antibodies. For instance, in drug development, IC50 values are used to assess the potency of inhibitors against enzymes or to neutralize ligand/receptor binding.
EC50 (Effective Concentration 50) represents the concentration of compound required to produce 50% of a desired effect (e.g., 50% of the maximum achievable response). It can be used to measure the potency of agonists such as a receptor ligand, hormone, or neurotransmitter. It is also used in pharmacology studies to determine the potency of a drug in eliciting a therapeutic effect.
How do you determine the IC50 of a compound?
It is determined by performing a dose-response analysis in which increasing concentrations of the compound are added to the cells (biological effect) or to the purified enzyme (biochemical effect) and the desired effect is quantified. The dose-response must be designed to capture potency, with lower concentration(s) showing lack of effect and high concentration(s) showing maximum effect.
- Design a serial dilution of the compound of interest in 3-fold increments. BPS Bioscience uses 9 to 10 concentrations in a dose-response.
- Include internal controls appropriate for the type of assay, and a control without compound. All assays require a “blank” (in the absence of cells or enzyme, signal originating from the reagents).
- Confirm that you are looking at the active enzyme or biological effect. In some cases, you may need to add an agonist to activate the cells or the enzyme.
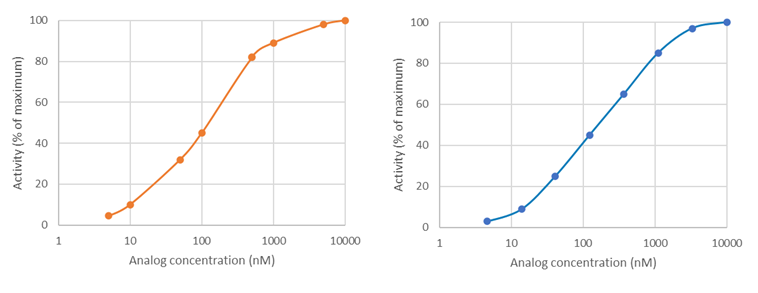
Why does BPS Bioscience recommend 3-fold dilutions to measure inhibitors IC50?
Most of the time, BPS Bioscience uses a base 10 Log scale for graphs, in which 3-fold dilutions are logarithmic and evenly spaced, whereas linear dilutions on a log scale may be clumped together. See these two theoretical plots: on the left, the compound concentrations were selected linearly and appear clumped by two. On the right, the compound concentrations were selected logarithmically and are evenly spaced. The logarithmic scale provides better sampling and is a better experimental design.
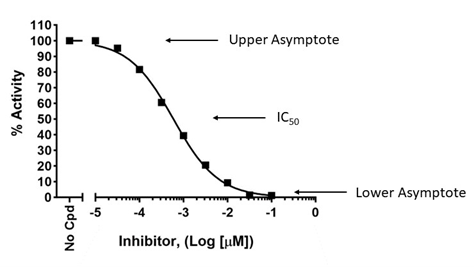
How do you know if you did not choose the correct concentration range?
To accurately determine an IC50, a dose-response must be designed to capture true potency, with lower concentration(s) showing lack of effect and high concentration(s) showing maximum effect. This allows to determine the higher and lower asymptotes on the graph, which are then used to calculate the position of the 50% value, as shown below (upper graph). If the higher concentrations do not cross the x-axis and do not reach a plateau, the true IC50 cannot be determined as the graph lacks the lower asymptote, as shown in the lower graph. Conversely, the lowest concentrations should exert no effect compared to the no-compound control.
To learn more, visit Sebaugh JL, Guidelines for accurate EC50/IC50 estimation
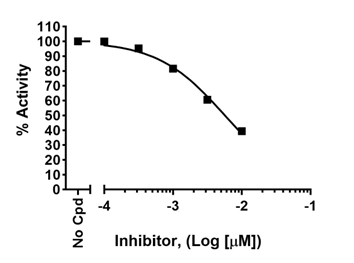
To learn more, visit Sebaugh JL, Guidelines for accurate EC50/IC50 estimation


Background issues: how do I design my colorimetric assay when my compound has color?
If the compound being tested has color that interferes with the assay (for example, a yellow compound would interfere with colorimetric assays read at λ = 570–590 nm), a blank must be determined for each concentration of the compound and the dose-response signal must be normalized against the blank for each compound concentration.
Why is the apparent IC50 value in the validation experiment different from the published value for this compound?
First, let’s define what “different” means for IC50 values. A 2 or 3-fold variation is usually considered within range and The IC50 would not be considered different.
Next, remember that IC50 values are not constants. They differ depending on the type of assay and the parameters of the assay. Assay sensitivity is one of the critical parameters affecting the measured IC50. Thus, IC50 values obtained using a colorimetric assay kit will not be similar to IC50 values obtained using Mass Spec. In addition, within the same assay format, enzyme to substrate ratio, time of reaction, and whether the compound is pre-incubated or not with the enzyme prior to reaction initiation, among other parameters, may affect the value obtained.
When comparing compound efficacy in drug discovery campaigns, it is critical to use the same assay format and the same experimental conditions for all compared compounds. Ideally, the enzyme used throughout the campaign should be standardized for specific activity.
Next, remember that IC50 values are not constants. They differ depending on the type of assay and the parameters of the assay. Assay sensitivity is one of the critical parameters affecting the measured IC50. Thus, IC50 values obtained using a colorimetric assay kit will not be similar to IC50 values obtained using Mass Spec. In addition, within the same assay format, enzyme to substrate ratio, time of reaction, and whether the compound is pre-incubated or not with the enzyme prior to reaction initiation, among other parameters, may affect the value obtained.
When comparing compound efficacy in drug discovery campaigns, it is critical to use the same assay format and the same experimental conditions for all compared compounds. Ideally, the enzyme used throughout the campaign should be standardized for specific activity.
What is a Fluorescence Polarization (FP) assay?
Fluorescence Polarization (FP) is based on measuring changes in light polarization emitted by a fluorescent probe in a sample. It is quite different from fluorescence intensity, which measures the intensity of emitted light at a specific wavelength. FP is widely used to monitor molecular interactions in solution. FP is a complex technique that requires careful design, involves somewhat complicated calculations, and uses a specific instrument. It is, therefore, important to understand the principles underlying FP technology and FP-based experiments. An overview of the technology, principles of FP-based experiments, advantages and considerations, can be found here [FP principles].
What type of instrument is needed?
A “standard” fluorescence plate reader measuring fluorescence intensity at a given wavelength will not capture FP data. The assay requires the use of an instrument capable of excitation with polarized light and capable of measuring fluorescence intensity on two different planes. For optimal experimental design, it is important to read the protocol provided with the kit all the way through before starting the experiment.
Why kind of assay plate is needed to perform an FP assay?
FP assays are performed in a black low-binding assay plate. Our kits include one assay plate per kit. The plate catalog number is indicated in the datasheet, should you need to purchase additional plates.
How sensitive is an FP assay?
FP technology measures fluorescence intensity emitted by the fluorophore in the two planes of light that are parallel and perpendicular relative to the plane of excitation. The degree of fluorescence polarization is defined as (P). Most instruments display fluorescence polarization in units of mP in which 1 mP = 1000 P.
Theoretical P values range from –0.33 to 0.5 (–330 to 500 mP), while experimental data typically range from 10 mP to around 300 mP.
FP instruments can achieve very precise measurements (± 2 mP).
Theoretical P values range from –0.33 to 0.5 (–330 to 500 mP), while experimental data typically range from 10 mP to around 300 mP.
FP instruments can achieve very precise measurements (± 2 mP).
Are the FP assay kits suitable for high throughput screening?
FP assay kits are ideal for high throughput screening because they are designed for small volumes and no wash steps are necessary. Most of our kits exist in a 96 well format, although some have also been optimized for use in a 384 well format.
What controls are needed to run a successful FP assay?
Several controls are needed to perform a successful FP assay. Controls are described in the protocol provided with BPS Bioscience assay kits.
For enzymatic reactions that require an agonist to work, keep in mind that the agonist should be added in all conditions except the negative control.
- Blank: the blank is meant to measure the background signal from the buffers used in the reaction. If studying a compound dissolved in an organic solvent, the same concentration of solvent should be present in all the controls. The Blank value is subtracted from all other values.
- Low FP: internal control, calibration for the lowest signal allowed by the particular assay being performed. For example, a condition in which all the fluorescent tracer is in free form.
- High FP: internal control, calibration for the highest signal allowed by the particular assay being performed. For example, a condition in which all the fluorescent tracer is in bound form.
- Experimental control: a condition lacking the compound under study. If the compound is an agonist, the “no compound” condition is the negative control. If the compound is an inhibitor, the “no compound” condition is the positive control, and this value should be set as 100% for calculations.
For enzymatic reactions that require an agonist to work, keep in mind that the agonist should be added in all conditions except the negative control.
Are FP assay kits suitable to use with crude biological matrices?
FP assays may allow the interrogation of complex solutions (for example a urine sample) as long as they are well defined and free of contaminants. However, the purity and quality of the samples are critical. Potential interference in light scattering can be caused by many large molecules such as cell or membrane debris. For this reason, crude cell lysates, cell culture supernatants and other rudimentary extracts should not be used in this type of assay. The presence of contaminants with high background fluorescence or with non-specific trapping ability is likely to result in high noise-to-signal ratios or to otherwise interfere with the signal.
The specificity of our kits is determined by the purified enzyme. If the sample contains any trace of another enzyme that can modify the substrate, the kit will not function properly as the trace enzymatic activity will interfere.
The specificity of our kits is determined by the purified enzyme. If the sample contains any trace of another enzyme that can modify the substrate, the kit will not function properly as the trace enzymatic activity will interfere.
Can I add BSA to my sample?
Bovine Serum Albumin (BSA) may interfere with some fluorescence kits. If BSA is needed, it is important to evaluate its effect by comparing polarization of the fluorescent tracer with or without BSA in the assay optimization phase.
A few assays developed by BPS bioscience contain BSA and have been optimized accordingly.
A few assays developed by BPS bioscience contain BSA and have been optimized accordingly.
What can cause high background fluorescence signals?
An unacceptably high background signal (provided by the “Blank”) can be caused by contaminants in the buffers. Attention to raw materials, cleanliness of mixing and storage vessels, and buffer preparation methods should reduce high background signals. High backgrounds can also be caused by solvents* that fluoresce at the wavelength of interest, which would be indicated by an unusually high signal in the “blank”. When using an uncommon diluent for the first time, it can be useful to compare the signal with and without diluent to assess any effect of the diluent on the background signal.
*Solvents: used to dissolve a compound that is not soluble in aqueous buffers. Typical organic solvents include DMSO, ethanol or dimethylfluoride.
*Solvents: used to dissolve a compound that is not soluble in aqueous buffers. Typical organic solvents include DMSO, ethanol or dimethylfluoride.
What are contraindications?
Contraindications are substances that interfere with the assay reaction. Anytime BPS Bioscience’s scientists observe that a solvent interferes with an assay, such as DMSO at a concentration of >1%, the observation will be indicated in the datasheet. Look for the “contraindications” section that will indicate known buffer or solvent interferences. Please keep in mind that BPS Bioscience does not test all solvents.
Some of the enzymatic assays can be impaired by specific buffers (for example phosphate-containing buffers interfere with Phosphodiesterase kits).
Some of the enzymatic assays can be impaired by specific buffers (for example phosphate-containing buffers interfere with Phosphodiesterase kits).
What is the “Gain” value setting?
The "Gain settings" are instrument-specific. Related information is usually found in the instrument owner’s manual. Most instruments have software programs that automatically adjust the gain settings based on inputs provided by the user when selecting the FP mode.
Typical settings:
Typical settings:
- Use the Blank subtraction setting. Select Blank reduction option for calculation of mP values. As a result, no value should appear in the Blank samples since the Blank is subtracted from all values obtained from the assay plate.
- Input the G factor. The G factor is instrument specific and can vary from instrument to instrument. The G factor is typically between 0.5-1.5, may be provided by the owner’s manual or may be automatically computed by the instrument. If manual input is required by the instrument, determine the G factor as detailed below.
- Select the low reference control. This will be the background signal when calculating the signal to background ratio.
- No negative mP values should be obtained. Negative values are obtained when the perpendicular fluorescence readings are greater than the parallel fluorescence readings, which is caused by incorrect settings.
What is the G factor?
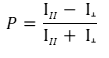
The P equation assumes that light is transmitted equally well through both parallel and perpendicular channels. In practice, this is generally not true and a correction must be made. This correction factor is called the "G Factor"; it is specific to the instrument and has to be determined by the user. Modern instruments usually have the G factor pre-calculated, so refer to your instrument instructions to determine how to set or calculate the G factor.
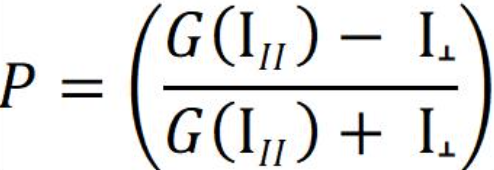
If you do need to calculate the G factor:
Use the theoretical mP of your fluorophore in the equation above (for example, the theoretical mP = 27 or P = 0.027 for fluorescein and Texas Red), together with the measurements for both the parallel and the perpendicular channels obtained from the free fluorophore at a concentration that gives counts well above the background “blank”. The background needs to be subtracted from both the parallel and the perpendicular fluorophore-only measurements. From the equation above, calculate:
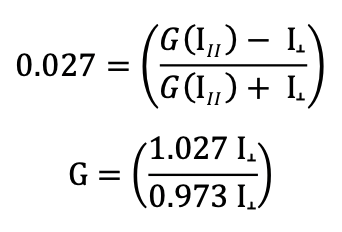
How do I determine the G factor when measuring FP?
The user may ignore the G-factor when all experiments are performed using the same instrument, since the G-factor is instrument-dependent.
Where needed, the G-factor should be set before measurements are performed. Some manuals will clearly say what the G-factor is. For example, our scientists use a Tecan fluorescent plate reader which has a G value set to 22 mP.
However, the G-factor may need to be figured out by the investigator when not clearly indicated by the manufacturer. The instrument manual will contain information about how to establish the G-factor.
Where needed, the G-factor should be set before measurements are performed. Some manuals will clearly say what the G-factor is. For example, our scientists use a Tecan fluorescent plate reader which has a G value set to 22 mP.
However, the G-factor may need to be figured out by the investigator when not clearly indicated by the manufacturer. The instrument manual will contain information about how to establish the G-factor.
How do I calculate Fluorescence Polarization results?
Instruments give measurements in milli-Polarization = mP.
Calculate ΔmP for all samples:
ΔmP = (mP value of the sample) – (mP value of the Reference control)
Where mP refers to milli-Polarization values provided by the instrument and Reference control is the mP value obtained in the condition containing only the fluorescent probe (a condition in which the probe is in free state).
Calculate ΔmP for all samples:
ΔmP = (mP value of the sample) – (mP value of the Reference control)
Where mP refers to milli-Polarization values provided by the instrument and Reference control is the mP value obtained in the condition containing only the fluorescent probe (a condition in which the probe is in free state).
What if the Alpha-counts signal of positive control reaction is same as “Blank” value?
Possible Causes and Solutions
Enzyme has lost activity: Enzyme loses activity upon re-peated freeze/thaw cycles. Use fresh JMJD2A, BPS Bioscience #50123. Store enzyme in single use aliquots. Increase time of enzyme incubation. Increase enzyme concentration.
Streptavidin Donor beads or anti-mIgG acceptor beads fail to show significant signal: Reorder Streptavidin Donor beads or anti-mIgG acceptor beads from Perkin Elmer.
Incorrect settings on instruments: Refer to instrument instructions for correct settings to increase sensitivity of light detection.
Enzyme has lost activity: Enzyme loses activity upon re-peated freeze/thaw cycles. Use fresh JMJD2A, BPS Bioscience #50123. Store enzyme in single use aliquots. Increase time of enzyme incubation. Increase enzyme concentration.
Streptavidin Donor beads or anti-mIgG acceptor beads fail to show significant signal: Reorder Streptavidin Donor beads or anti-mIgG acceptor beads from Perkin Elmer.
Incorrect settings on instruments: Refer to instrument instructions for correct settings to increase sensitivity of light detection.
The Alpha-counts signal is erratic or varies widely among wells. What could be wrong?
Inaccurate pipetting/technique: Run duplicates of all reactions. Use a multichannel pipettor. Use master mixes to minimize errors.
What is a TR-FRET assay?
Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) is commonly used to analyze the binding of two interacting molecules. The technology is a combination of Time-Resolved Fluorescence and Förster’s Resonance Energy Transfer (FRET), a phenomenon in which a light-excited fluorophore can transfer its absorbed energy to a nearby acceptor fluorophore. An eBook describing the principle of TR-FRET and examples is available to download or print [eBooks].
What are the advantages of TR-FRET experiments?
TR-FRET is an ultra-low background technique allowing the measure of any reaction in which two labeled entities come in proximity. The main drawbacks of the technique are that it requires two optimized labeled entities, in addition to exhibiting a low dynamic range. However, these drawbacks are offset by several advantages:

- Simple protocols, fast assays
- Small volumes
- Homogeneous: no need for washing steps or for physical separation from the unbound entities
- Robust, sensitive signal
- Ultra-low background with high signal to noise ratio
- Stable signal: use of lanthanide donor fluorophores minimizes photobleaching
What is Förster’s Resonance Energy Transfer (FRET)?
FRET is a phenomenon in which two fluorophores emitting at different wavelengths are coupled: the donor fluorophore excited by a high energy source transfers energy (not light) to an acceptor fluorophore. This results in excitation of the acceptor and fluorescence emission at another wavelength, which can be detected by a fluorescence reader.
The transfer of energy between the donor and the acceptor depends on physical proximity (<10 nm) and decreases rapidly with distance. Thus, partner molecules distributed in a solution are sufficiently far apart and FRET does not occur. Upon interaction, the partners complete the FRET pairing as they are now in proximity to each other.
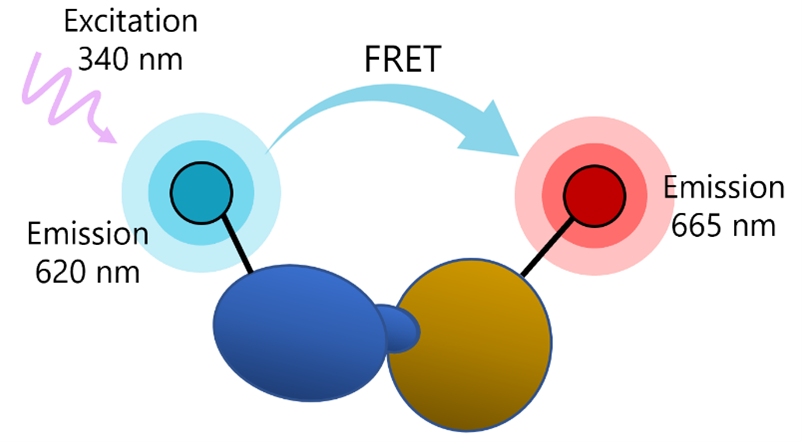
The transfer of energy between the donor and the acceptor depends on physical proximity (<10 nm) and decreases rapidly with distance. Thus, partner molecules distributed in a solution are sufficiently far apart and FRET does not occur. Upon interaction, the partners complete the FRET pairing as they are now in proximity to each other.

What does Time-Resolved Fluorescence (TRF) mean?
Classic fluorescence intensity uses short-lived fluorophores such as fluorescein, with an emission speed in the order of the nanoseconds. Excitation and emission occur at specific wavelengths that can be differentiated by a fluorescence reader. However, excitation and emission happen at the same time. If there is any amount of spectral overlap between excitation and emission, as there usually is, the reader will capture some of the excitation fluorescence, resulting in background signal and low signal-to-noise ratios.
TRF solves this by using long-lived inorganic fluorophores as donors and adding a time delay between excitation and measurement, which means that the excitation signal is gone by the time of the measurement, which decreases background signals. TRF also uses excitation pulses (not continuous excitation), so that a series of measurements are repeated over time. TRF also eliminates transient background fluorescence generated from sample components such as buffers, proteins, and chemicals, which hinders classic FRET methods. Thus, TRF technology accounts for the ultra-low background advantage of TR-FRET.
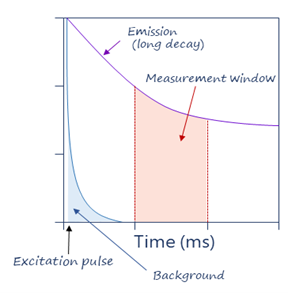
TRF solves this by using long-lived inorganic fluorophores as donors and adding a time delay between excitation and measurement, which means that the excitation signal is gone by the time of the measurement, which decreases background signals. TRF also uses excitation pulses (not continuous excitation), so that a series of measurements are repeated over time. TRF also eliminates transient background fluorescence generated from sample components such as buffers, proteins, and chemicals, which hinders classic FRET methods. Thus, TRF technology accounts for the ultra-low background advantage of TR-FRET.

What are the most common fluorophores?
Ideal fluorophores have high signal intensity, are highly stable, and offer excellent signal-to-noise ratios. Fluorophores commonly used are “Lanthanide probes” which are metal ions referring to elements Cerium to Lutetium in the periodic table. The most used are Europium and Terbium, which fluoresce over milliseconds instead of nanoseconds.
What type of instrument is needed?
A fluorescent plate reader capable of measuring Time Resolved-Fluorescence Resonance Energy Transfer is needed for these experiments.
How is measurement made?
In practice, a comparison measurement of the two emitted wavelengths over time is calculated for a TR-FRET response.
Therefore, two sequential measurements are conducted. For example, Tb-donor emission should be measured at 620 nm followed by dye-acceptor emission at 665 nm. Data analysis is performed using the TR-FRET ratio (665 nm emission/620 nm emission).
To calculate the percentage activity, subtract the Blank value from all other FRET values. It is expected that Blank and Negative Control have a similar value. The FRET value from the Positive Control can be set as one hundred percent activity as it corresponds to the maximum activity.
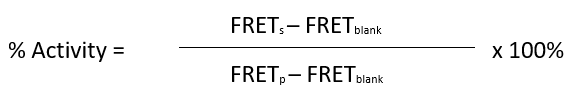
Where FRETs = Sample FRET, FRETblank = Blank FRET, and FRETP = Positive control FRET.
Therefore, two sequential measurements are conducted. For example, Tb-donor emission should be measured at 620 nm followed by dye-acceptor emission at 665 nm. Data analysis is performed using the TR-FRET ratio (665 nm emission/620 nm emission).
To calculate the percentage activity, subtract the Blank value from all other FRET values. It is expected that Blank and Negative Control have a similar value. The FRET value from the Positive Control can be set as one hundred percent activity as it corresponds to the maximum activity.
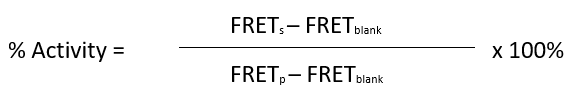
Where FRETs = Sample FRET, FRETblank = Blank FRET, and FRETP = Positive control FRET.
Why do I obtain a low luminescence signal in my positive control compared to my test inhibitor when using Kinase-Glo® MAX reagent in your kinase assay kits?
Kinases use ATP to add a phosphate group onto a substrate protein. The phosphorylation reaction, therefore, depletes ATP and generates ADP.
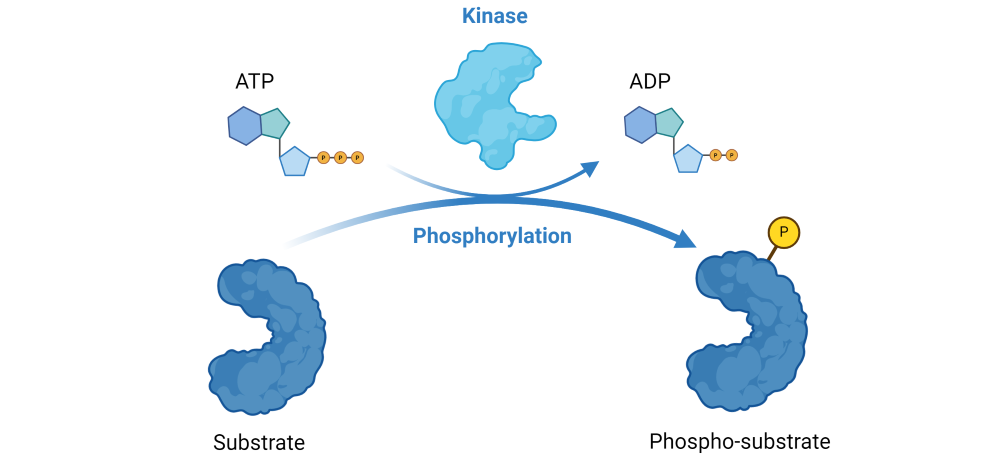
Figure 1: Illustration of the phosphorylation reaction catalyzed by kinases.
The Kinase-Glo® MAX reagent is added as a detection reagent that quantitatively measures the remaining, unused ATP. The reagent is linear up to 500 µM ATP.
Since the luminescent signal correlates with the amount of remaining ATP, it is inversely proportional to kinase activity. Thus, a decrease in the luminescent signal corresponds to higher kinase activity, while an increase in the signal corresponds to lower kinase activity.
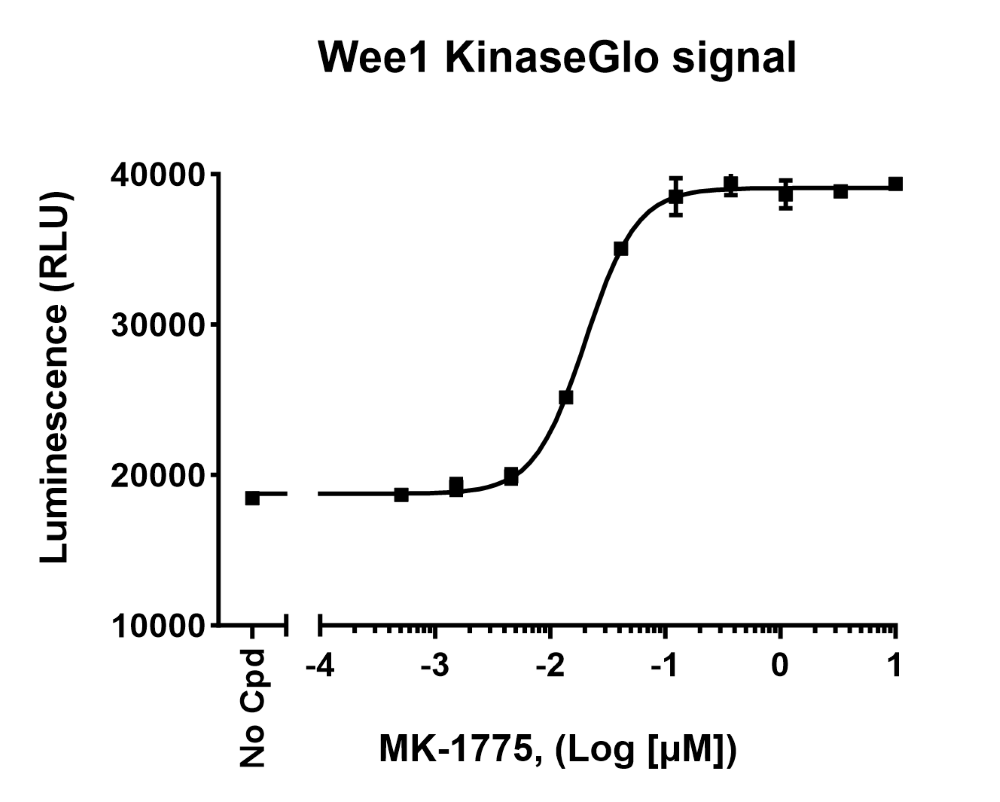
Figure 2: Raw luminescence data obtained by measuring Wee1 kinase activity in the presence of increasing concentrations of inhibitor MK-155, using Kinase-Glo® MAX reagent (Wee1 Kinase Assay Kit (#79909)).
Validation graphs shown in the assay kit datasheets show results expressed as percent of kinase activity, in which kinase activity in the absence of inhibitor (“positive control”) is set to 100%.
Results are calculated by averaging replicates and subtracting the blank from all experimental values. Thus, for each “test inhibitor” concentration, calculate:
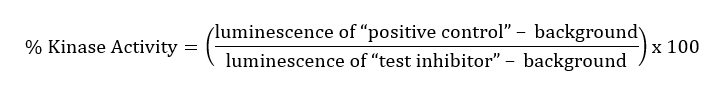
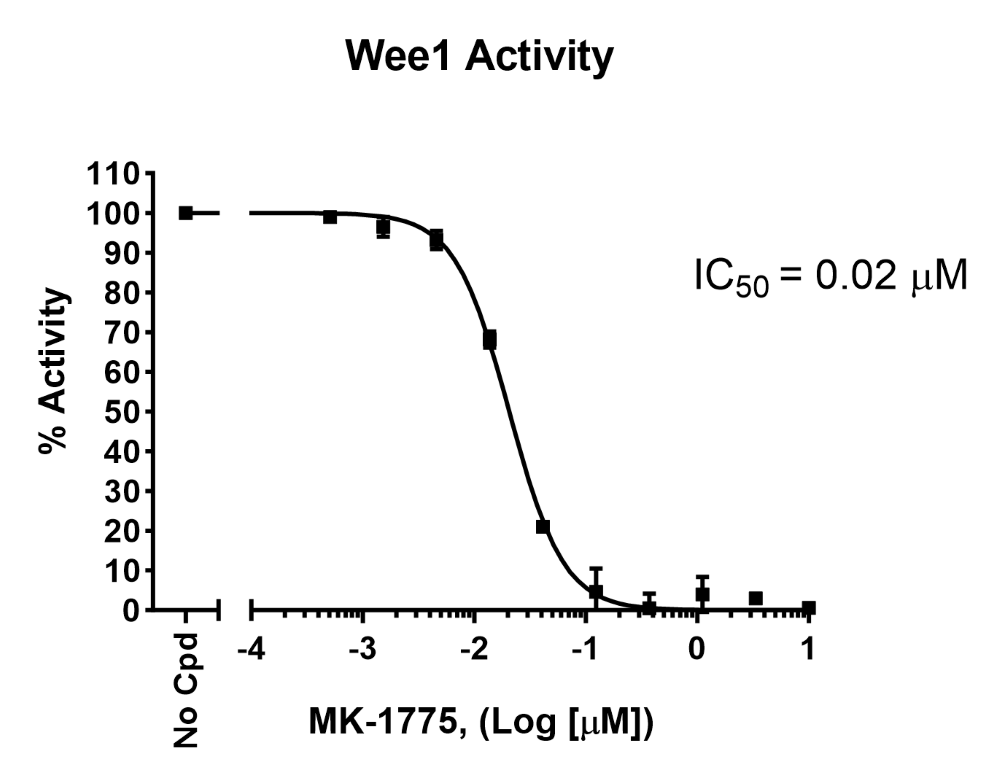
Figure 3: Inhibition of Wee1 kinase activity by MK-1775 measured using Wee1 Kinase Assay Kit (#79909). Results are expressed as percent of activity (positive control without MK-1775 set to 100%).

Figure 1: Illustration of the phosphorylation reaction catalyzed by kinases.
The Kinase-Glo® MAX reagent is added as a detection reagent that quantitatively measures the remaining, unused ATP. The reagent is linear up to 500 µM ATP.
Since the luminescent signal correlates with the amount of remaining ATP, it is inversely proportional to kinase activity. Thus, a decrease in the luminescent signal corresponds to higher kinase activity, while an increase in the signal corresponds to lower kinase activity.

Figure 2: Raw luminescence data obtained by measuring Wee1 kinase activity in the presence of increasing concentrations of inhibitor MK-155, using Kinase-Glo® MAX reagent (Wee1 Kinase Assay Kit (#79909)).
Validation graphs shown in the assay kit datasheets show results expressed as percent of kinase activity, in which kinase activity in the absence of inhibitor (“positive control”) is set to 100%.
Results are calculated by averaging replicates and subtracting the blank from all experimental values. Thus, for each “test inhibitor” concentration, calculate:
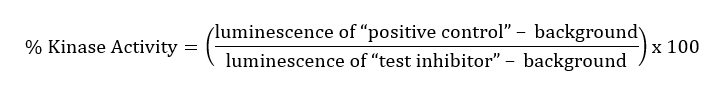

Figure 3: Inhibition of Wee1 kinase activity by MK-1775 measured using Wee1 Kinase Assay Kit (#79909). Results are expressed as percent of activity (positive control without MK-1775 set to 100%).

