PARP/PARPTrap™ Screening & Profiling
PARP (Poly ADP-Ribose Polymerase) proteins are a large family of 17 members that catalyze the ADP-ribosylation of proteins and DNA. PARPs are part of a network of 150 proteins involved in the DNA damage response, which constantly scan and repair DNA to maintain genome integrity. The PARP proteins are involved in a wide range of biological functions: DNA repair, chromatin remodeling, mitotic spindle assembly, regulation of RNA turnover, regulation of gene expression, apoptosis, and more.
Accelerate your drug discovery and development projects using PARP or PARPtrap™ screening services. We offer the largest PARP proteins panel on the market, allowing you to compare efficacy across our entire PARP family.
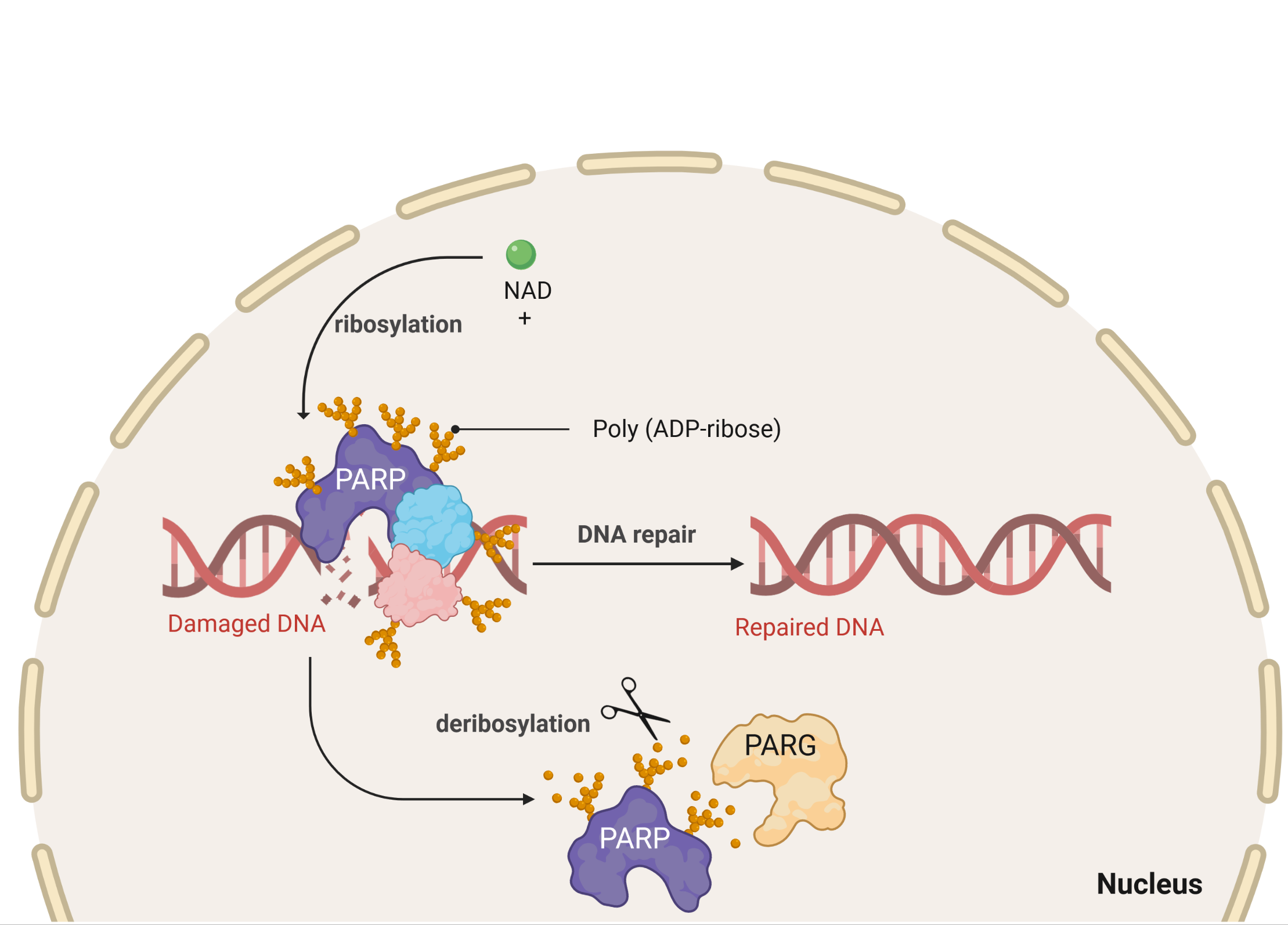
Our team of experts, using our in-house developed biochemical assay kits, will:
- Screen for PARP inhibitors from large compound libraries, measured by ELISA, AlphaLISA®, or Fluorescence Polarization assays
- Screen for PARG inhibitors
- Compare potency with IC50 screens
- Answer your questions and guide your project in a timely manner
- Deliver detailed results promptly and on time
If follow-up studies are required, we can provide the same active proteins as the ones used in our assays.
Learn more about the process and deliverables.
Enzymatic Activity by ELISA
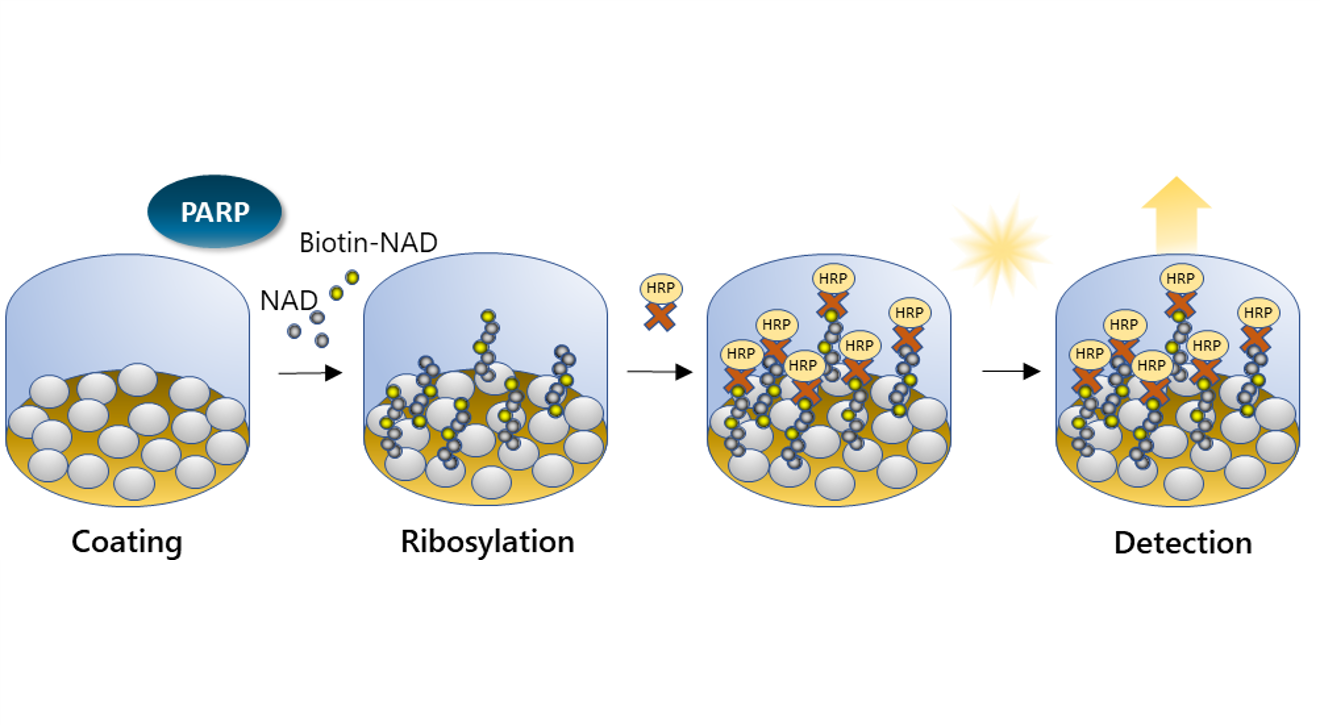
ELISA: Histone proteins are coated on a plate. Biotin-labeled NAD+ is added with the PARP enzyme. PARP-mediated ribosylation activity is detected by adding Streptavidin-HRP (horseradish peroxidase), which binds to the newly formed ribosylation branch, and a chemiluminescent or colorimetric HRP substrate.
Enzymatic Activity by AlphaLISA®
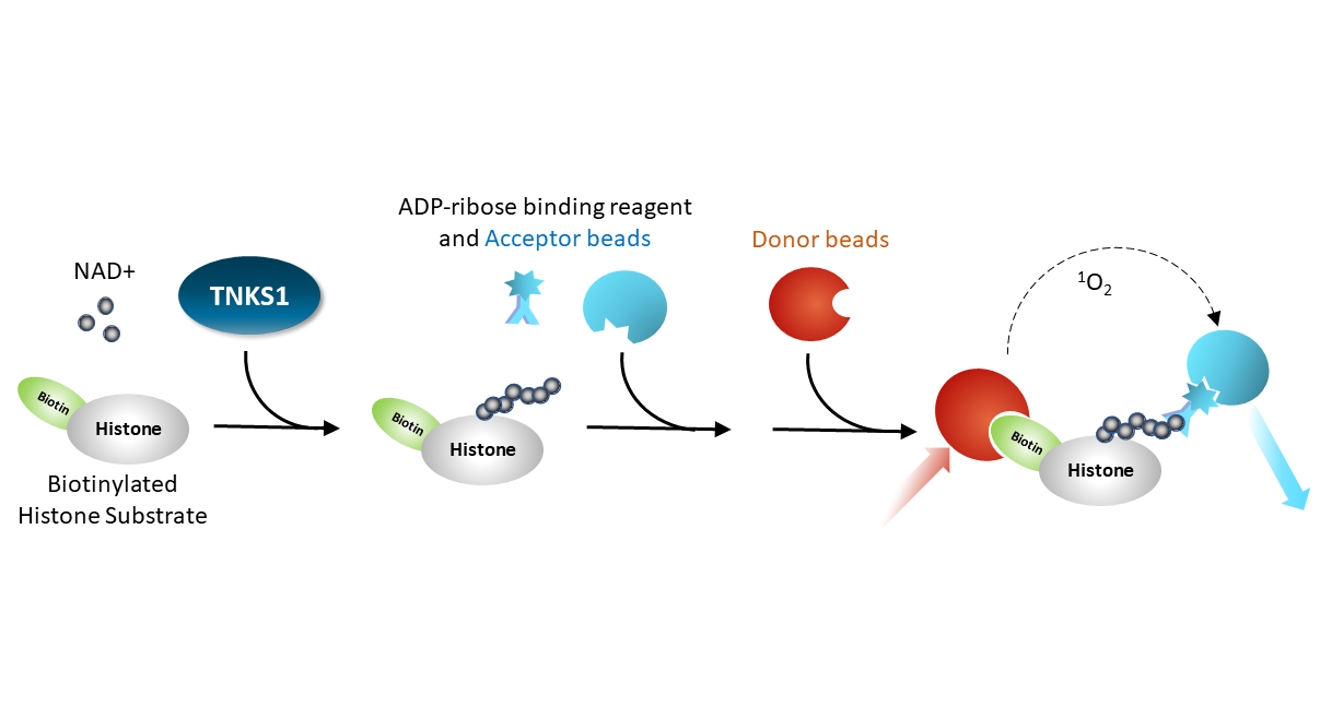
AlphaLISA® Assay: a PARP enzyme is incubated with biotinylated histone substrate and NAD+ for an hour. An ADP-ribose binding reagent is added with an acceptor bead, then a streptavidin-conjugated donor bead is added. Excitation of the donor bead results in excitation of the acceptor bead and light emission. This is a homogeneous assay.
PARP Trapping
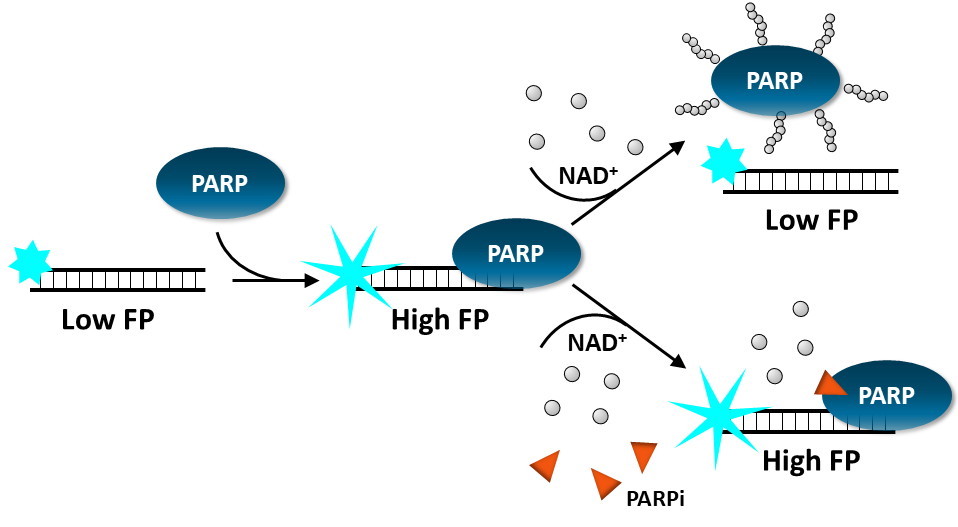
Fluorescence Polarization Assay: The assay uses fluorescent DNA probes that emit differently polarized light depending on PARP binding. Addition of a PARP inhibitor prevents ribosylation and results in the trapping of PARP onto the fluorescent oligonucleotide duplex, which increases the Fluorescence Polarization signal in a dose dependent manner. This is a homogeneous assay.
Molecular Degrader Optimization
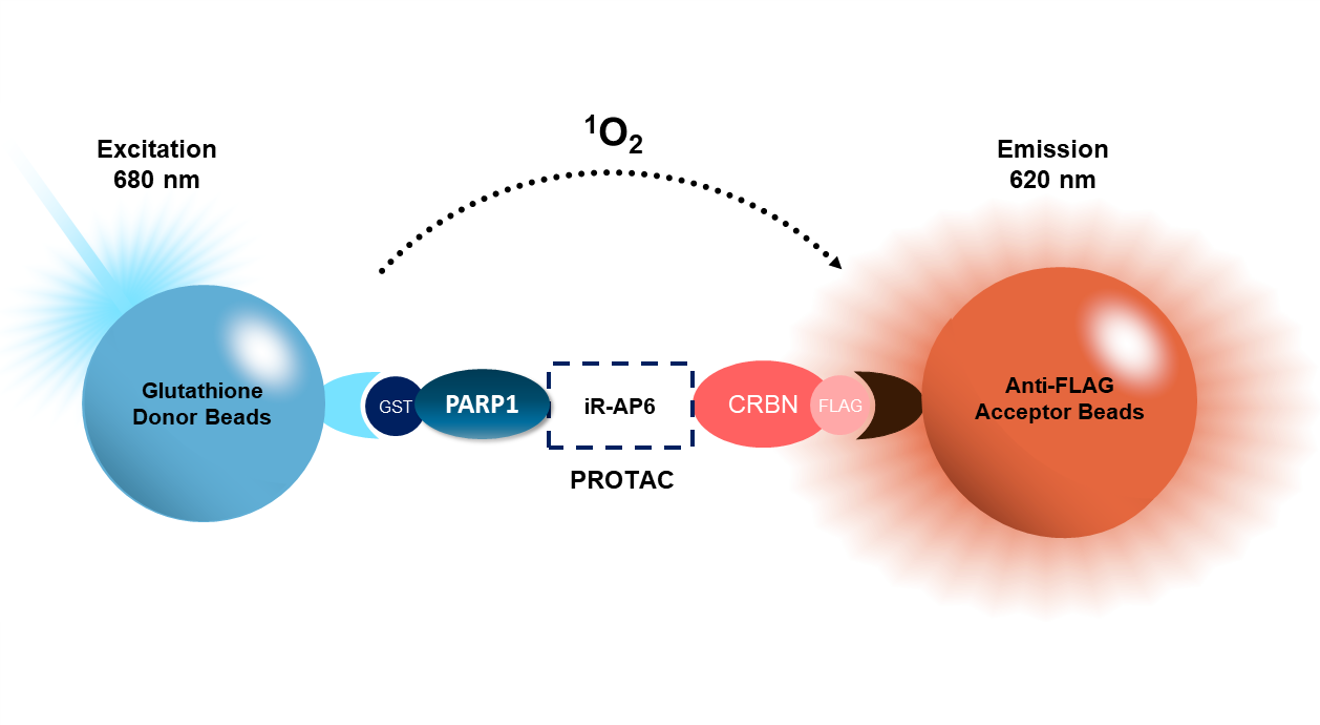
AlphaLISA® Assay: A degrader of interest interacts with both PARP and CRBN, bringing them in proximity. PARP-GST is recognized by the GSH donor bead, while CRBN-FLAG binds to anti-FLAG conjugated to the acceptor bead. Excitation of the donor bead results in excitation of the acceptor bead and light emission. This is a homogeneous assay.
Available Service Assays
Note: Not all targets will have a commercially available reference control. Please inquire with our team if you need to furnish your own controls for specific targets.

