Research Applications for Engineered iPS Cells
Introduction
New avenues of research were dramatically opened by the discovery of Takahashi and Yamanaka in 2006 that the ectopic expression of Oct4 (octamer-binding transcription factor 4), Sox2 (SRY-box2), Kfl4 (Kruppel-like factor 4) and c-Myc was sufficient to reprogram terminally differentiated cells into induced pluripotent stem (iPS) cells. These have infinite expansion potential and similar genetic markers, epigenetics and multilineage differentiation ability as ESCs (embryonic stem cells) (1). While ESC-based research requires the use of human blastocysts, iPS cells can be made from an adult skin biopsy, thus relieving stem cell research from some of its most controversial ethical concerns. iPSC can then be used in a variety of research and clinical applications.
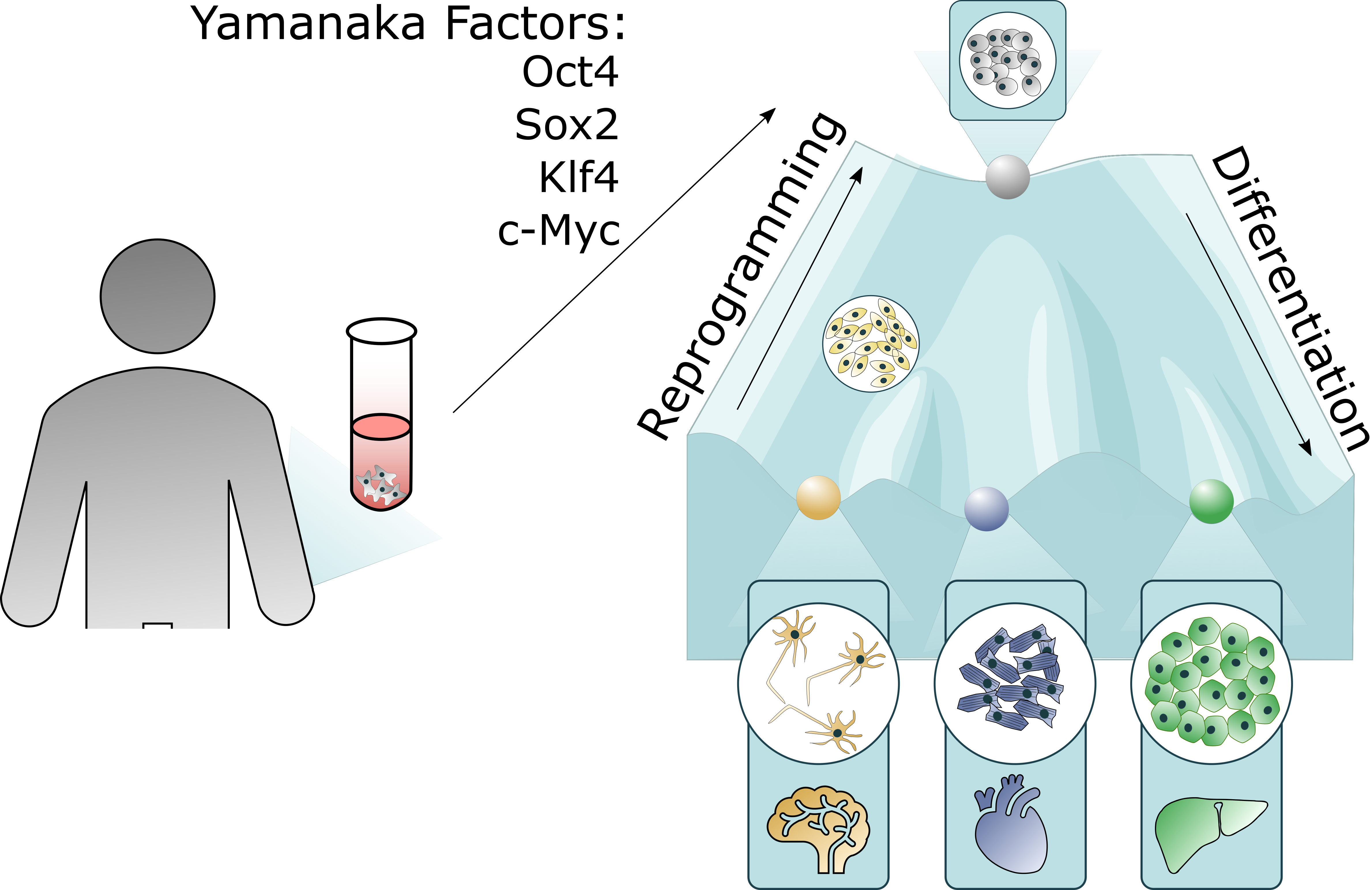
Figure 1: iPSC reprogramming and differentiation are inverse paths. The use of Yamanaka factors is sufficient to reprogram differentiated cells into iPSCs, which can then be driven to differentiate along specific pathways.
iPS cell as research models
The ability to reset the clock of differentiated cells to become stem cells, combined with the discovery of differentiation factors and conditions to specific lineages had a tremendous impact in biomedical research. iPSCs have been used successfully in disease modeling, drug discovery and toxicology screening.
Disease modeling
In the past, disease modeling studies mostly used overexpression/knockout/knockdown of known players in a disease, often in cell lines derived from a different tissue or that had lost most of their relevant properties. Those studies tended to be complemented, when possible, with post-mortem tissue analysis from patients. Researchers were then combining information from a cell system that no longer resembled the model it wanted to mimic, with one single time point analysis that captured only the catastrophic result of the pathology and had obliterated all the clues that could be used to stop disease progression at an early stage. Animals were the nearest models to the human body, but often were not close enough to reflect human pathology. In addition, concerns with animal welfare and reduction in live animal research are now very important considerations.
Using iPSC, we are now able to collect tissue biopsies of healthy individuals and/or with specific diseases and mutations to mimic the disease and its progression in a dish, deepening our physiological and pathological knowledge. For example, the use of iPS-derived cardiomyocytes from a patient with hypoplastic left heart syndrome (HLHS), a congenital heart disease, allowed researchers to identify deficiencies in differentiation, sarcomere organization, and slower contraction, as responsible for the clinical outcome (2). Another field that has strongly benefited from the “disease in a dish” approach is neuroscience. In the past neurological diseases studies were often conducted in non-nervous system cell lines but today we can model Alzheimer’s disease (AD), Parkinson’s disease (PD), amyloid lateral sclerosis (AL), Huntington’s disease, Machado-Joseph disease and even schizophrenia. iPS cells can faithfully reproduce aspects of disease, such as the accumulation of alpha-synuclein in iPS-derived neural cells from a patient with PA, while allowing identification of cellular dysfunctions previously unknown (deficient axonal transport, neurite extension and retraction, etc) (3).
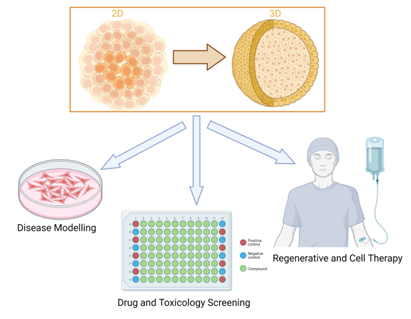
Figure 2: iPS cells can be used as 2D or 3D models in disease modeling, drug and toxicity screening and regenerative medicine and cell therapy.
This new understanding of pathologies opens the door to new treatment options, prior to the irreversible onset of disease.
Modeling diseases with iPSC has been a driving force in the development of many new or improved technologies but has also strongly benefited from the advances in genome sequencing and engineering tools. This combination has brought hope to patients who had little. Rare diseases often remained unstudied since the investment into creating cell models and study them was seen as too high versus the profit that could be generated. Or we simply did not have the ability to create complex cellular models that could be used. Today, by combining iPS technology with whole genome sequencing, mutations can be easily identified. We can then make use of technologies like CRISPR/Cas9 and create iPS lines where the mutation can be introduced or corrected. The direct link between genotype and phenotype can be made in a fairly efficient and quick way. One example where these advances have been highly beneficial is retinal studies. The retina is composed of multiple highly specialized interconnected cell types, such as the RPE (retinal pigment epithelium) and PR (photoreceptors). These are terminally differentiated cells prone to ageing related degeneration and to mutational impact. While in the past we were unable to mimic such a high level of complexity, we can now generate RPE and even mini retinas that allow us to understand tissue development, pathways and impact of mutations.
Drug discovery
Another highly relevant application of iPS cells is in drug development and screening. With iPSC we have gained deeper understanding of the molecular pathways involved in a disease, found new phenotypes, and are now also able to generate relevant cellular models, in enough number, to perform high throughput screening (HTS) with easy read outs, such as luciferase or GFP expression. For example, iPS derived motor neurons from ALS patients allowed the screening of a large library of compounds able to suppress neuron cell death and identified a drug already in use for the treatment of chronic myelogenous leukemia, bosutinib (3). We can even look back at current treatment options and try to understand their real benefit in light of our newly acquired knowledge of the disease. This is the case in schizophrenia, where iPSC derived from schizophrenic patients have shown lower levels of PSD95 (postsynaptic density protein-95), glutamate receptor, and decreased axon number. Testing of five commonly used antipsychotic drugs indicated that only one had a real effect in increasing neural connectivity.
In addition to drug development, toxicity and cell permeability studies can now also be conducted in relevant cell systems. The ability to test toxicity profiles of drug candidates in cell types that can mimic more closely the actual target tissue, and even its complexity, can increase the predictability of drug behavior in humans. A better understanding of the toxicology profile of a candidate drug can benefit the patients by identifying drugs that can act with fewer severe side effects, while avoiding large and expensive dose escalation studies.
iPS cells as therapeutic tools
Initial studies had focused on the generation of autologous cell therapies, where the patient’s cells would be reprogrammed, genetic mutations would be corrected (if applicable), differentiated into the cell type needed and reintroduced into the patient. While this is still possible in certain cases, it has become clear that the timeframe involved is not compatible with the progression of some diseases. Attention has been shifting to the use of allogeneic alternatives, and the creation of ‘off-the-shelf” therapies. The use of allogeneic cells brings its own concerns, such as the potential for immune rejection due to the mismatch of HLAs (human leukocyte antigens) between donor and recipient. Countries like Japan are attempting to create a bank of iPS cells that covers 95% of the Japanese population, a major undertaking that will require more than 140 different cell lines. Other strategies involve the creation of universal donor lines, where B2M (beta 2 microglobulin) and CIITA (class II major histocompatibility complex transactivator) are not present and thus do not express class I and II HLA on the cell surface. Clinical trials based on iPS derived cells are underway in the US, China, Germany, and Japan, targeting a number of diseases (examples can be seen in Table 1) (1,3).
Table 1: Examples of diseases targeted in clinical trials using iPS derived cells, and cell types being used.
| Disease | Cell Type |
|---|---|
| AMD (Age Macular Degeneration) | RPE (Retinal Pigment Epithelium) |
| RP (Retinitis Pigmentosa) | Retinal sheet |
| Limbal Stem Cell Deficiency | Limbal Epithelial Stem Cells |
| Parkinson’s Disease | Dopamine Neural Progenitor Cells |
| Spinal cord injuries | Neural Progenitor Cells |
| Heart Failure | Cardiomyocytes |
| Aplastic Anemia | Platelets |
| Thrombocytopenia | Megakaryocytes |
| Advanced Head and Neck Cancer | iNKT cells |
| Articular Cartilage Damage | Chondrocytes |
| Urea Cycle Disorder | Hepatocytes |
| Chronic Kidney Disease | Nephron Progenitor Cells |
| Recessive Dystrophic Epidermolysis Bullosa | Keratinocytes |
| Multiple Sclerosis | Oligodendrocyte Precursor Cells |
While bringing major advantages, certain aspects should be kept in mind. Interestingly, iPSCs tend to retain some epigenetic memory, which impacts their ability to differentiate (4), and should undergo strict quality control to determine the presence of tumorigenic cells in the final cell population, including the residual presence of iPSCs. Another aspect often neglected is the need to assess the cell maturity level, via specific markers and characteristics. The use of immature cells can result in detrimental consequences, as in the case of cell replacement with immature cardiomyocytes which generate arrhythmia.
The advent of iPSC derived organoids
The idea that a 2D cellular system does not represent closely the complexity of an organ has always been present in researchers’ minds. 3D spheroids represented a step towards a complex system for HTS drug screening. However, these are unable to mimic a tissue’s complexity as they lack organization and cell type heterogeneity. On the other hand, stem cells have the intrinsic ability to assemble into complex structures when cultured and can form organoids. Organoids are 3D self-organized tissue models that recapitulate spatial organization of heterogeneous tissue-specific cells, cell-cell, cell-matrix interactions, while allowing extended culture and manipulation. Examples of organoids include optic cup, brain, intestine, liver and kidney (6). These can be used in a wide range of applications.
The development of organoids needs to be aligned with specific applications. When considering developmental studies, the focus may be on mimicking as close as possible the in vivo environment, while for regenerative medicine the major effort is put into scalability. Drug screening applications require a recapitulation of only critical aspects of the in vivo tissue that are relevant to the testing and create an easy readout (5). The introduction of reporter markers, such as luciferase, can provide a quick and well-established readout method, while technologies such as encapsulation in microspheres and organ-on-a-chip allow HTS. Organoids were found to have a high predictive value in drug responses, and represent a major step forward in drug development.
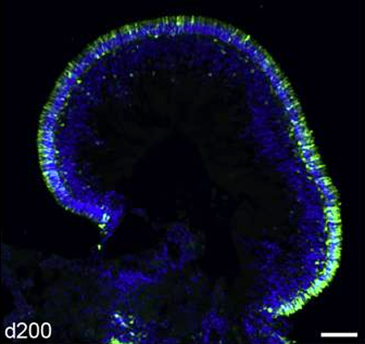
Figure 3: Retina organoid derived from iPS cells using GFP as reporter. Fluorescent image of a cryosection of human retina organoid developed in vitro for 200 days, with GFP being expressed under the control of the CRX (cone-rod homeobox) promoter as photoreceptor marker. Green = GFP positive cells; Blue = DAPI staining of cell nuclei (7).
Conclusions
The future is bright for iPS cell-based research and therapy. Fine tuning of aspects such as our ability to model late onset diseases in a short timeframe, develop vascularization and specific spatial-temporal differentiation control, will advance the field and allow better therapies. BPS Bioscience offers iPS cell lines expressing reporter genes, such as GFP and luciferase, that allow an easy readout in HTS drug screening and toxicity assays. In addition, we also provide Cas9 expressing iPS cells, a perfect tool for efficient genetic manipulation for your studies of disease in a dish or therapeutic applications, making us an ideal partner in your research endeavors.
References
(1) Okano H. and Yamanaka S., 2014 Mol Brain 7:22.
(2) Grskovi M., et al., 2011, Nature Reviews Drug Discovery 10: 915-929.
(3) Tsujimoto H and Osafune K., 2022 FEBS 289 (23):7274-7291.
(4) Yuasa S., et al., 2022 Front Cell Dev Biol, 9: doi.org/10.3389/fcell.2021.831304.
(5) Yin X., et al., 2016 Cell Stem Cell. 18(1):25-38.
(6) Ye L., et al., 2013 Curr Cardiol Rev 9(1): 63-72
(7) Kruczek K., et al., 2021 Stem Cell Reports 16 (2): 252-263.

