Lentiviruses for SARS-CoV-2 Research
To understand the mechanism of SARS-CoV-2 cell entry, it is essential to study how Spike proteins interact with the ACE2 receptor. However, such studies are hampered by the danger of producing and manipulating live coronavirus. A safer alternative to using live coronavirus is to use recombinant lentivirus pseudotyped with the coronavirus S protein. The Spike pseudotyped lentivirus includes a luciferase reporter, allowing the monitoring of viral entry into host cells. BPS also offers the control (bald) lentivirus without Spike protein as well as ACE2 lentivirus to transduce cells for transient expression or to create stable cell lines as targets for the Spike pseudovirion. BPS offers a variety of SARS-CoV-2 related lentiviruses.
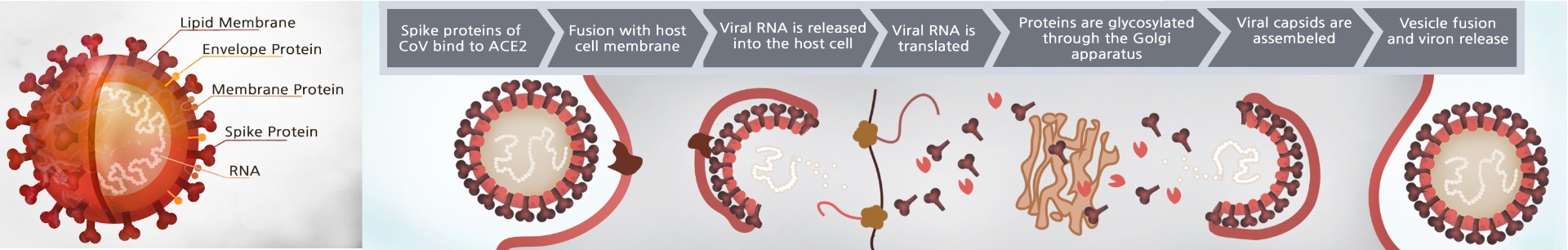
SARS-CoV-2 interacts and penetrates into human cells through the binding of the Spike protein localized out of it’s membrane and ACE2 (angiotensin converting enzyme 2), a homodimer receptor found on the surface of some human cells, particularly those in the respiratory tract. The Spike protein, which consists of two subunit (S1 and S2), promotes attachment to ACE2 thanks to the subunit S1 and its RBD (Receptor Binding Domain) region. After enzymatic cleavage by cellular proteases (furin and TMPRSS2) the S2 subunit facilitates fusion of the viral and cellular membrane allowing the virion to enter the cell. Based on this observation, drugs targeting the interaction between the Spike protein of SARS-CoV-2 and ACE2 may offer protection against the viral infection.
BPS Bioscience is actively working to provide COVID-19 research tools to support scientists in their work to find an efficient treatment. These research tools include antibodies, assay kits, lentiviruses, proteins, and substrates. The BPS Bioscience biochemical assays are used to identify inhibitors of the Spike–ACE2 interaction. Based on the sandwich ELISA technique, these 96 well plate assays allow quick screening and identification of compounds (chemicals or antibodies) able to block the Spike:ACE2 interaction and virus penetration.
Variant Lentiviruses
Spike (B.1.351 Variant) (SARS-CoV-2) Pseudotyped Lentivirus (Luc Reporter)
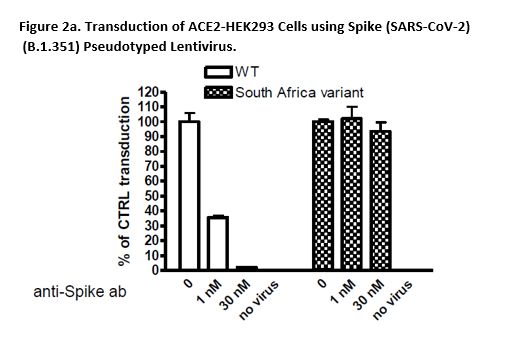 A variant called B.1.351 was first identified in the fall of 2020 in the Republic of South Africa. This South African variant, also known as 501Y.V2, has many mutations which may lead to higher transmissibility and infectivity. The Spike (B.1.351 Variant) (SARS-CoV-2) Pseudotyped Lentivirus were produced with SARS-CoV-2 B.1.351 Variant Spike (Genbank Accession #QHD43416.1 with B.1.351 mutations; as the envelope glycoproteins instead of the commonly used VSV-G.
A variant called B.1.351 was first identified in the fall of 2020 in the Republic of South Africa. This South African variant, also known as 501Y.V2, has many mutations which may lead to higher transmissibility and infectivity. The Spike (B.1.351 Variant) (SARS-CoV-2) Pseudotyped Lentivirus were produced with SARS-CoV-2 B.1.351 Variant Spike (Genbank Accession #QHD43416.1 with B.1.351 mutations; as the envelope glycoproteins instead of the commonly used VSV-G.
These pseudovirions contain the firefly luciferase gene driven by a CMV promoter, therefore, the spike-mediated cell entry can be measured via luciferase activity. The Spike (B.1.351 Variant) (SARS-CoV-2) pseudotyped lentivirus can be used to measure the activity of neutralizing antibody against SARS-CoV-2 B.1.351 variant in a Biosafety Level 2 facility.
Spike (P.1 Variant) (SARS-CoV-2) Pseudotyped Lentivirus (Luc Reporter)
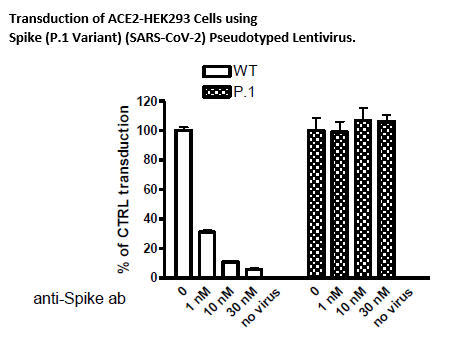 In Brazil, a variant called P.1 was first identified in the summer of 2020. This variant has many mutations that may lead to higher transmissibility and infectivity. The Spike (P.1) (SARS-CoV-2) Pseudotyped Lentiviruses were produced with SARS-CoV-2 Variant Spike (Genbank Accession #QHD43416.1 with P.1 mutations, see below for details) as the envelope glycoproteins instead of the commonly used VSVG.
In Brazil, a variant called P.1 was first identified in the summer of 2020. This variant has many mutations that may lead to higher transmissibility and infectivity. The Spike (P.1) (SARS-CoV-2) Pseudotyped Lentiviruses were produced with SARS-CoV-2 Variant Spike (Genbank Accession #QHD43416.1 with P.1 mutations, see below for details) as the envelope glycoproteins instead of the commonly used VSVG.
These pseudovirions contain the firefly luciferase gene driven by a CMV promoter, therefore, the spike-mediated cell entry can be measured via luciferase activity. The Spike (P.1) (SARS-CoV-2) pseudotyped lentivirus can be used to measure the activity of neutralizing antibody against SARS-CoV-2 (P.1) variant using a Biosafety Level 2 facility.
Spike (K417T, E484K, N501Y) (SARS-CoV-2) Pseudotyped Lentivirus (Luc Reporter)
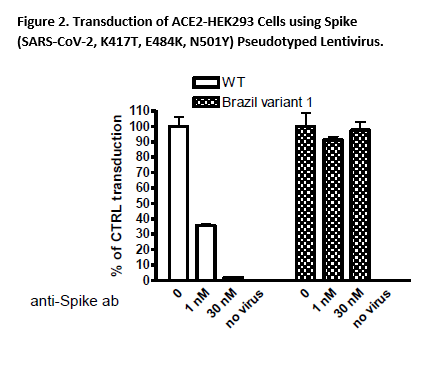 In Brazil, a variant called B.1.1.28.1 (P.1) was first identified in the fall of 2020. This variant has many mutations which may lead to higher transmissibility and infectivity. Among these mutations, three (K417T, E484K, N501Y) appear to be crucial. The Spike (K417T, E484K, N501Y) (SARS-CoV-2) Pseudotyped Lentiviruses were produced with SARS-CoV-2 Variant Spike (Genbank Accession #QHD43416.1 with mutations K417T, E484K, and N501Y) as the envelope glycoproteins instead of the commonly used VSV-G.
In Brazil, a variant called B.1.1.28.1 (P.1) was first identified in the fall of 2020. This variant has many mutations which may lead to higher transmissibility and infectivity. Among these mutations, three (K417T, E484K, N501Y) appear to be crucial. The Spike (K417T, E484K, N501Y) (SARS-CoV-2) Pseudotyped Lentiviruses were produced with SARS-CoV-2 Variant Spike (Genbank Accession #QHD43416.1 with mutations K417T, E484K, and N501Y) as the envelope glycoproteins instead of the commonly used VSV-G.
These pseudovirions contain the firefly luciferase gene driven by a CMV promoter, therefore, the spike-mediated cell entry can be measured via luciferase activity. The Spike (SARS-CoV-2, K417T, E484K, N501Y) pseudotyped lentivirus can be used to measure the activity of neutralizing antibody against SARS-CoV-2 K417T, E484K, N501Y variant in intact cells using a Biosafety Level 2 facility.
Spike (SARS-CoV-2, B.1.1.7) Pseudotyped Lentivirus (Luc Reporter)
Spike (SARS-CoV-2, D614G) Pseudotyped Lentivirus (eGFP Reporter)
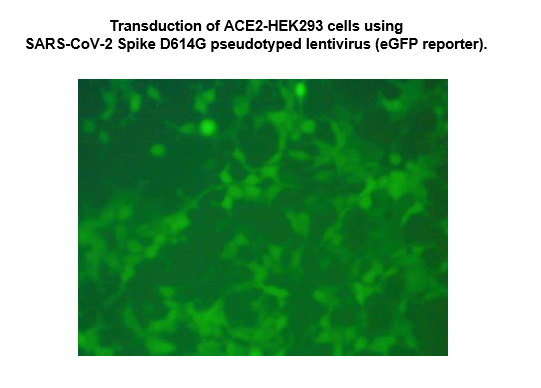 The SARS-CoV-2 Spike D614G Pseudotyped Lentivirus were produced with SARS-CoV-2 Spike (Genbank Accession #QHD43416.1; with D614G mutation) as the envelope glycoproteins instead of the commonly used VSV-G. These pseudovirions contain the enhanced GFP gene driven by a CMV promoter, therefore, the spike-mediated cell entry can be conveniently determined via eGFP activity. The SARS-CoV-2 Spike D614G pseudotyped lentivirus can be used to measure the activity of neutralizing antibody against SARS-CoV-2 in a Biosafety Level 2 facility.
The SARS-CoV-2 Spike D614G Pseudotyped Lentivirus were produced with SARS-CoV-2 Spike (Genbank Accession #QHD43416.1; with D614G mutation) as the envelope glycoproteins instead of the commonly used VSV-G. These pseudovirions contain the enhanced GFP gene driven by a CMV promoter, therefore, the spike-mediated cell entry can be conveniently determined via eGFP activity. The SARS-CoV-2 Spike D614G pseudotyped lentivirus can be used to measure the activity of neutralizing antibody against SARS-CoV-2 in a Biosafety Level 2 facility.
Spike (SARS-CoV-2, D614G) Pseudotyped Lentivirus (Luc Reporter)
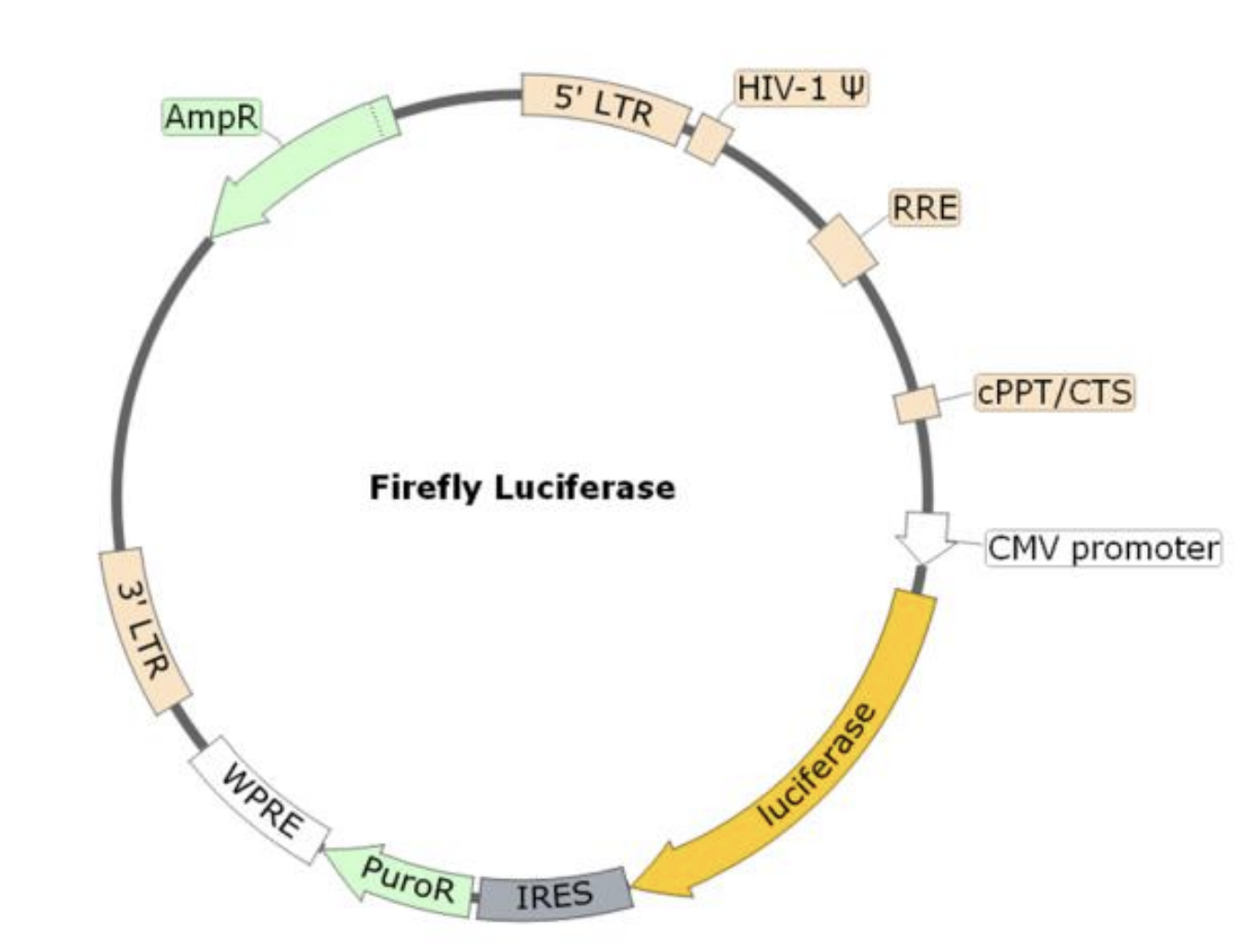 The SARS-CoV-2 Spike D614G Pseudotyped Lentivirus were produced with SARS-CoV-2 Spike (Genbank Accession #QHD43416.1; with D614G mutation) as the envelope glycoproteins instead of the commonly used VSV-G. These pseudovirions contain the firefly luciferase gene driven by a CMV promoter, therefore, the spike-mediated cell entry can be measured via luciferase activity. The SARS-CoV-2 Spike D614G pseudotyped lentivirus can be used to measure the activity of neutralizing antibody against SARS-CoV-2 in a Biosafety Level 2 facility.
The SARS-CoV-2 Spike D614G Pseudotyped Lentivirus were produced with SARS-CoV-2 Spike (Genbank Accession #QHD43416.1; with D614G mutation) as the envelope glycoproteins instead of the commonly used VSV-G. These pseudovirions contain the firefly luciferase gene driven by a CMV promoter, therefore, the spike-mediated cell entry can be measured via luciferase activity. The SARS-CoV-2 Spike D614G pseudotyped lentivirus can be used to measure the activity of neutralizing antibody against SARS-CoV-2 in a Biosafety Level 2 facility.
Spike Reporter Lentiviruses
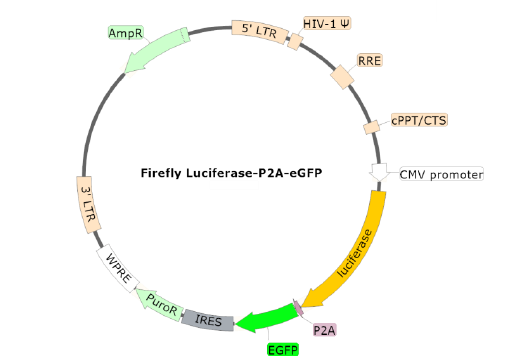
Spike(SARS-CoV-2) Pseudotyped Lentivirus (Luc-eGFP Dual Reporter)
Spike (SARS-CoV-2) Pseudotyped Lentivirus (Luciferase Reporter)
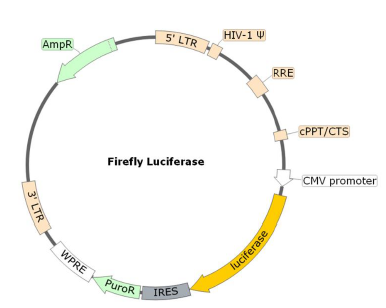
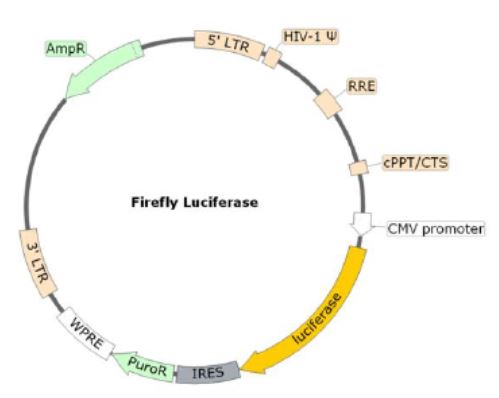
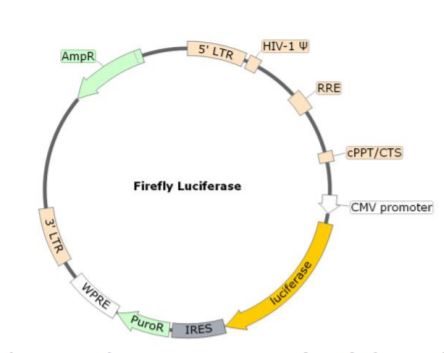 Due to its interaction with the Human ACE2, the coronavirus Spike protein is involved in the first step of the viral replication, which is the attachment of the virus to the host cell. In combination with the ACE2 lentivirus, BPS Bioscience has developed the Spike (SARS-CoV-2) Pseudotyped Lentivirus. Here, the commonly used VSV-G viral fusion protein, which classically mediates the fusion between the virus envelope and host cellular membrane, has been replaced by the SARS-CoV-2 Spike protein (Genbank Accession #QHD43416.1) as the envelope glycoproteins. These pseudovirions also contain the firefly luciferase gene driven by a CMV promoter, to easily measure the spike-mediated cell entry via luciferase reporter activity. Luciferase detection was measured with the ONE-Step Luciferase reagent. Lentivirus Figure: Schematic of the Luciferase Reporter in SARS-CoV-2 Spike Pseudovirion.
Due to its interaction with the Human ACE2, the coronavirus Spike protein is involved in the first step of the viral replication, which is the attachment of the virus to the host cell. In combination with the ACE2 lentivirus, BPS Bioscience has developed the Spike (SARS-CoV-2) Pseudotyped Lentivirus. Here, the commonly used VSV-G viral fusion protein, which classically mediates the fusion between the virus envelope and host cellular membrane, has been replaced by the SARS-CoV-2 Spike protein (Genbank Accession #QHD43416.1) as the envelope glycoproteins. These pseudovirions also contain the firefly luciferase gene driven by a CMV promoter, to easily measure the spike-mediated cell entry via luciferase reporter activity. Luciferase detection was measured with the ONE-Step Luciferase reagent. Lentivirus Figure: Schematic of the Luciferase Reporter in SARS-CoV-2 Spike Pseudovirion.
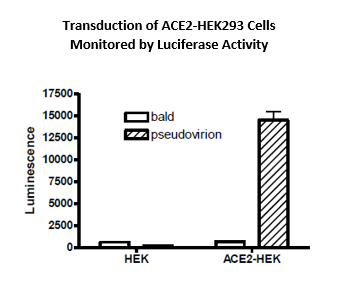
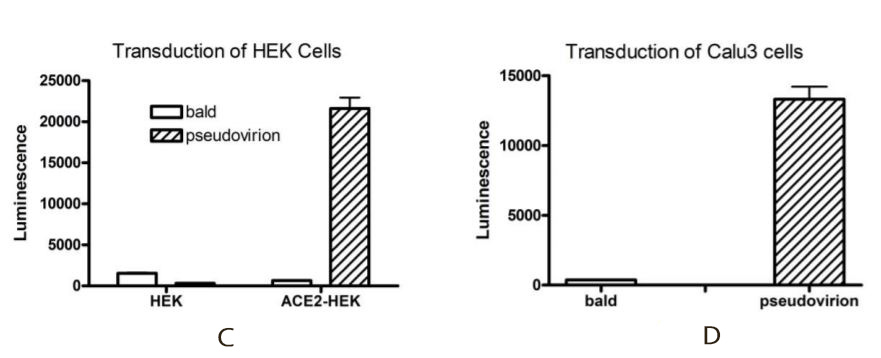 Transduction efficiency of the Spike (SARS-CoV-2) pseudotyped lentivirus has been evaluated on HEK293 cells expressing or not expressing the ACE2 protein (figure C) and on Calu3 cells (human lung cancer cell line which are epithelial and can act as respiratory models in preclinical applications) (figure D) used as a transduction control. Based on luciferase measurements, it appears that the Spike (SARS-CoV-2) pseudotyped lentivirus transduced ACE2-HEK293 and Calu3 cells with much greater efficiency compared with HEK293 parental cells. It indicates that the transduction is dependent upon ACE2 expression. The bald lentiviral pseudovirion, where no envelope glycoprotein is expressed, was used as a negative control.
Transduction efficiency of the Spike (SARS-CoV-2) pseudotyped lentivirus has been evaluated on HEK293 cells expressing or not expressing the ACE2 protein (figure C) and on Calu3 cells (human lung cancer cell line which are epithelial and can act as respiratory models in preclinical applications) (figure D) used as a transduction control. Based on luciferase measurements, it appears that the Spike (SARS-CoV-2) pseudotyped lentivirus transduced ACE2-HEK293 and Calu3 cells with much greater efficiency compared with HEK293 parental cells. It indicates that the transduction is dependent upon ACE2 expression. The bald lentiviral pseudovirion, where no envelope glycoprotein is expressed, was used as a negative control.
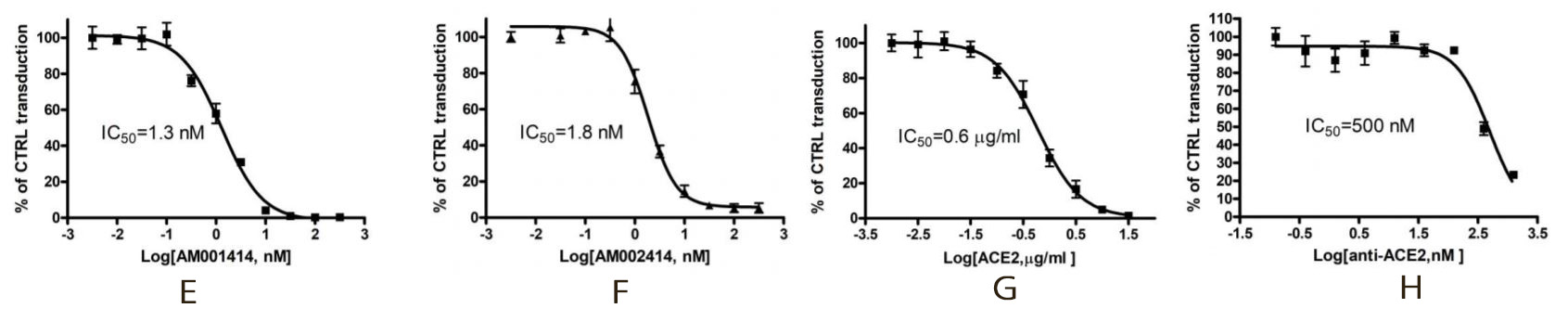
With ACE2 dependent transduction efficiency proved, several neutralization assays have been performed with an anti-SARS-CoV-2 Spike antibody (figures E and F), a recombinant ACE2 protein (figure G) and an anti-ACE2 antibody (figure H).
For the three neutralizing assays, the HEK293 cells expressing ACE2, were transduced with a mix containing the Spike (SARS-CoV-2) pseudotyped lentivirus and different concentrations of the corresponding neutralizing compounds. The percentage of control transduction corresponds to the luciferase measured in the control wells with the ACE2 expressing cells and the virus, without any neutralizing compounds. These three independent experiments showed a lower transduction efficiency of the Spike (SARS-CoV-2) Pseudotyped Lentivirus (decrease in the luciferase expression), due to an inhibitory effect of the tested compounds.
In combination with ACE2 Lentivirus, this Spike (SARS-CoV-2) pseudotyped lentivirus can be used to study the mechanism of viral transduction, and easily screen for neutralizing compounds (antibodies – chemical compounds) of the SARS-CoV-2 Spike and ACE2 interaction, in a Biosafety Level 2 facility.
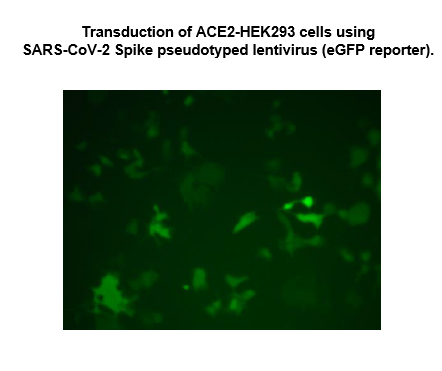
Spike (SARS-CoV-2) Pseudotyped Lentivirus (eGFP Reporter)
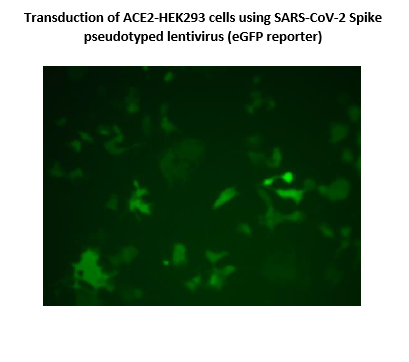 The SARS-CoV-2 Spike Pseudotyped Lentivirus were produced with SARS-CoV-2 Spike (Genbank Accession #QHD43416.1) as the envelope glycoproteins instead of the commonly used VSV-G. These pseudovirions also contain the enhanced GFP gene driven by a CMV promoter (Figure 1), therefore, the spike-mediated cell entry can be conveniently determined via eGFP activity. The SARS-CoV-2 Spike pseudotyped lentivirus can be used to measure the activity of neutralizing antibody against SARS-CoV-2 in a Biosafety Level 2 facility.
The SARS-CoV-2 Spike Pseudotyped Lentivirus were produced with SARS-CoV-2 Spike (Genbank Accession #QHD43416.1) as the envelope glycoproteins instead of the commonly used VSV-G. These pseudovirions also contain the enhanced GFP gene driven by a CMV promoter (Figure 1), therefore, the spike-mediated cell entry can be conveniently determined via eGFP activity. The SARS-CoV-2 Spike pseudotyped lentivirus can be used to measure the activity of neutralizing antibody against SARS-CoV-2 in a Biosafety Level 2 facility.
VSVg Pseudotyped Lentiviruses
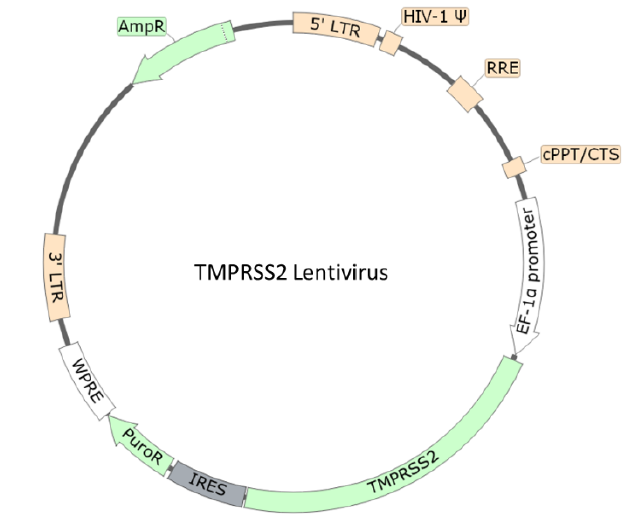
TMPRSS2 Lentivirus
Human transmembrane serine protease 2 (TMPRSS2) is an enzyme primarily expressed by endothelial cells across the respiratory and digestive tracts. It is involved in viral entry and spread of coronaviruses including SARS-CoV-2, the virus that causes COVID19. TMPRSS2 primes the spike protein by cleaving at the S1/S2 site, leading to the virus fusing to the respiratory epithelia on the cell surface through binding to ACE2. Blocking TMPRSS2 activity could potentially be an effective clinical therapy for COVID-19.
The TMPRSS2 Lentivirus are replication incompetent, HIV-based, VSV-G pseudotyped lentiviral particles that are ready to be transduced into almost all types of mammalian cells, including primary and non-dividing cells. The particles contain an TMPRSS2 gene (NM_005656.4) driven by an EF1a promoter.
Spike (SARS-CoV-2) Lentivirus
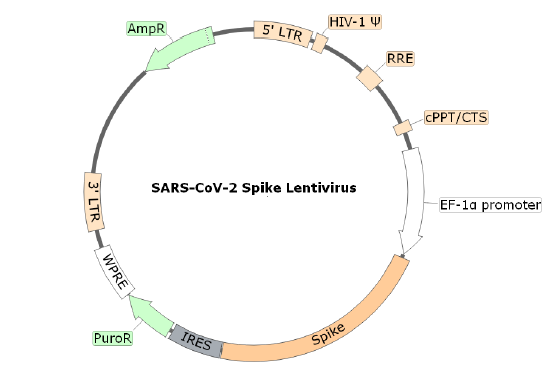
Cell entry of SARS-CoV-2 depends on the binding of viral spike protein to cellular receptor ACE2. The SARS-CoV-2 Spike Lentivirus are replication incompetent, HIV-based, VSV-G pseudotyped lentiviral particles that are ready to be transduced into almost all types mammalian cells, including primary and non-dividing cells. The particles contain the full length SARS-CoV-2 spike gene (QHD43416.1) driven by an EF1a promoter .
ACE2 Lentivirus
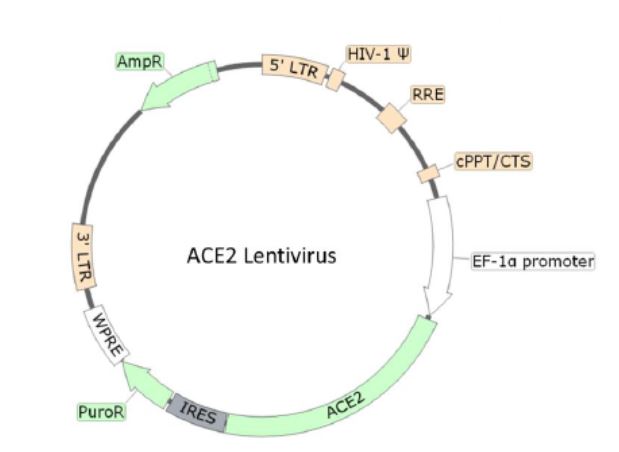 Localized on the surface of many human cell types (lungs, arteries, heart, kidney, and intestines), ACE2 is the entry point of the virus due to its interaction with the coronavirus Spike membrane protein. The ACE2 Lentivirus is replication incompetent, HIV-based, VSV-G pseudotyped lentiviral particles that are ready to be transduced into almost all types of mammalian cells, including primary and non-dividing cells. Under the control of EF1a promoter, the particles contain an ACE2 gene (NM_021804.3) allowing a transient expression of ACE2 in your target cell or a generation of a stable cell line expressing ACE2 with Puromycin.
Localized on the surface of many human cell types (lungs, arteries, heart, kidney, and intestines), ACE2 is the entry point of the virus due to its interaction with the coronavirus Spike membrane protein. The ACE2 Lentivirus is replication incompetent, HIV-based, VSV-G pseudotyped lentiviral particles that are ready to be transduced into almost all types of mammalian cells, including primary and non-dividing cells. Under the control of EF1a promoter, the particles contain an ACE2 gene (NM_021804.3) allowing a transient expression of ACE2 in your target cell or a generation of a stable cell line expressing ACE2 with Puromycin.
Lentivirus Figure: Schematic of the lenti-vector used to generate the ACE2 lentivirus
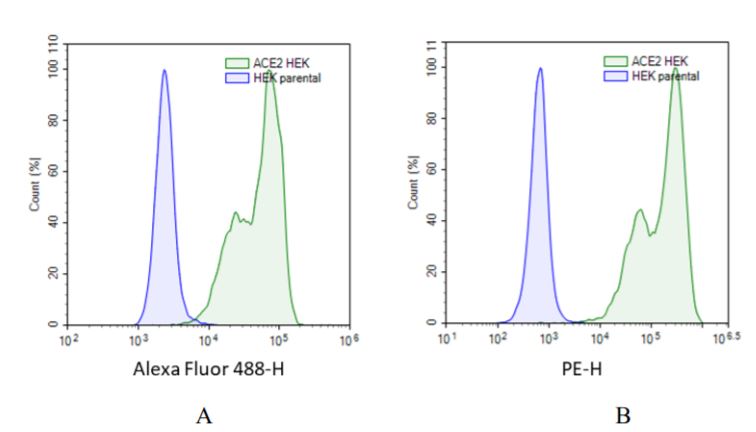
The transduction efficiency and the expression of ACE2 in HEK293 cells has been evaluated by FACS analysis with two different strategies. The first used a specific anti-human ACE2 antibody (figure A). The transduced cells were stained by anti-human ACE2 polyclonal goat IgG primary antibody and Alexa Fluor 488-conjugated rabbit anti-goat IgG as a secondary antibody. In the second strategies (figure B), the transduced cells were stained by a specific ACE2 ligand (biotinylated Spike S1 protein) and the Phycoerythrin (PE) conjugated to Streptavidin for the detection. In both cases FACS analysis reveal a high level of ACE2 expression at the surface of the HEK293 cells.
Figures A & B: FACS analysis of the ACE2 expression in HEK293 cells transduced with ACE2 lentivirus. Blue: HEK293 parental cells; Green: HEK293 cells transduced with ACE2 lentivirus.
Bald Lentiviruses
Bald Lentiviral Pseudovirion (Luciferase Reporter)
 The bald lentiviral pseudovirion was produced without envelope glycoproteins such as VSV-G or SARS-CoV-2 spike. It contains the firefly luciferase gene driven by a CMV promoter as the reporter. The bald lentiviral pseudovirion can serve as a negative control when studying virus entry initiated by specific interactions between virus particles and receptors.
The bald lentiviral pseudovirion was produced without envelope glycoproteins such as VSV-G or SARS-CoV-2 spike. It contains the firefly luciferase gene driven by a CMV promoter as the reporter. The bald lentiviral pseudovirion can serve as a negative control when studying virus entry initiated by specific interactions between virus particles and receptors.
Bald Lentiviral Pseudovirion (Luc-eGFP Dual Reporter)
 The bald lentiviral pseudovirion was produced without envelope glycoproteins such as VSV-G or SARS-CoV-2 spike. It contains a firefly luciferase and eGFP cassette (Luc-P2A-eGFP) as the reporters, driven by a CMV promoter. The bald lentiviral pseudovirion can serve as a negative control when studying virus entry initiated by specific interactions between virus particles and receptors.
The bald lentiviral pseudovirion was produced without envelope glycoproteins such as VSV-G or SARS-CoV-2 spike. It contains a firefly luciferase and eGFP cassette (Luc-P2A-eGFP) as the reporters, driven by a CMV promoter. The bald lentiviral pseudovirion can serve as a negative control when studying virus entry initiated by specific interactions between virus particles and receptors.
Bald Lentiviral Pseudovirion (eGFP Reporter)
 The bald lentiviral pseudovirion was produced without envelope glycoproteins such as VSV-G or SARS-CoV-2 spike. It contains the eGFP gene driven by a CMV promoter as the reporter. The bald lentiviral pseudovirion can serve as a negative control when studying virus entry initiated by specific interactions between virus particles and receptors.
The bald lentiviral pseudovirion was produced without envelope glycoproteins such as VSV-G or SARS-CoV-2 spike. It contains the eGFP gene driven by a CMV promoter as the reporter. The bald lentiviral pseudovirion can serve as a negative control when studying virus entry initiated by specific interactions between virus particles and receptors.
New Cell Signaling Lentiviruses
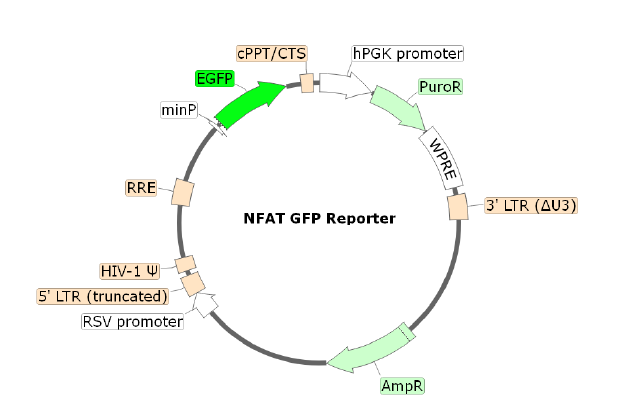
NFAT eGFP Reporter Lentivirus
YFP (Topaz) Lentivirus
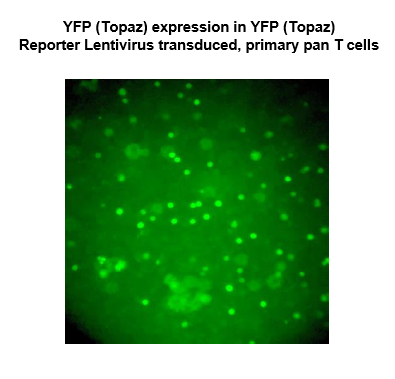 Topaz1, a yellow fluorescence protein (YFP), is one of the brightest and thermally stable green fluorescence proteins (GFP). The YFP (Topaz) Reporter Lentivirus contains replication-defective, VSV-G pseudotyped lentiviral particles that are ready to be transduced into almost all types of mammalian cells, including primary and non-dividing cells. The particles contain an EF1α promoter-driven YFP(Topaz) construct (a.a. sequence below), and confer both YFP (Topaz) expression and puromycin resistance to the target cells.
Topaz1, a yellow fluorescence protein (YFP), is one of the brightest and thermally stable green fluorescence proteins (GFP). The YFP (Topaz) Reporter Lentivirus contains replication-defective, VSV-G pseudotyped lentiviral particles that are ready to be transduced into almost all types of mammalian cells, including primary and non-dividing cells. The particles contain an EF1α promoter-driven YFP(Topaz) construct (a.a. sequence below), and confer both YFP (Topaz) expression and puromycin resistance to the target cells.