Enzymatic Biotinylation & BirA
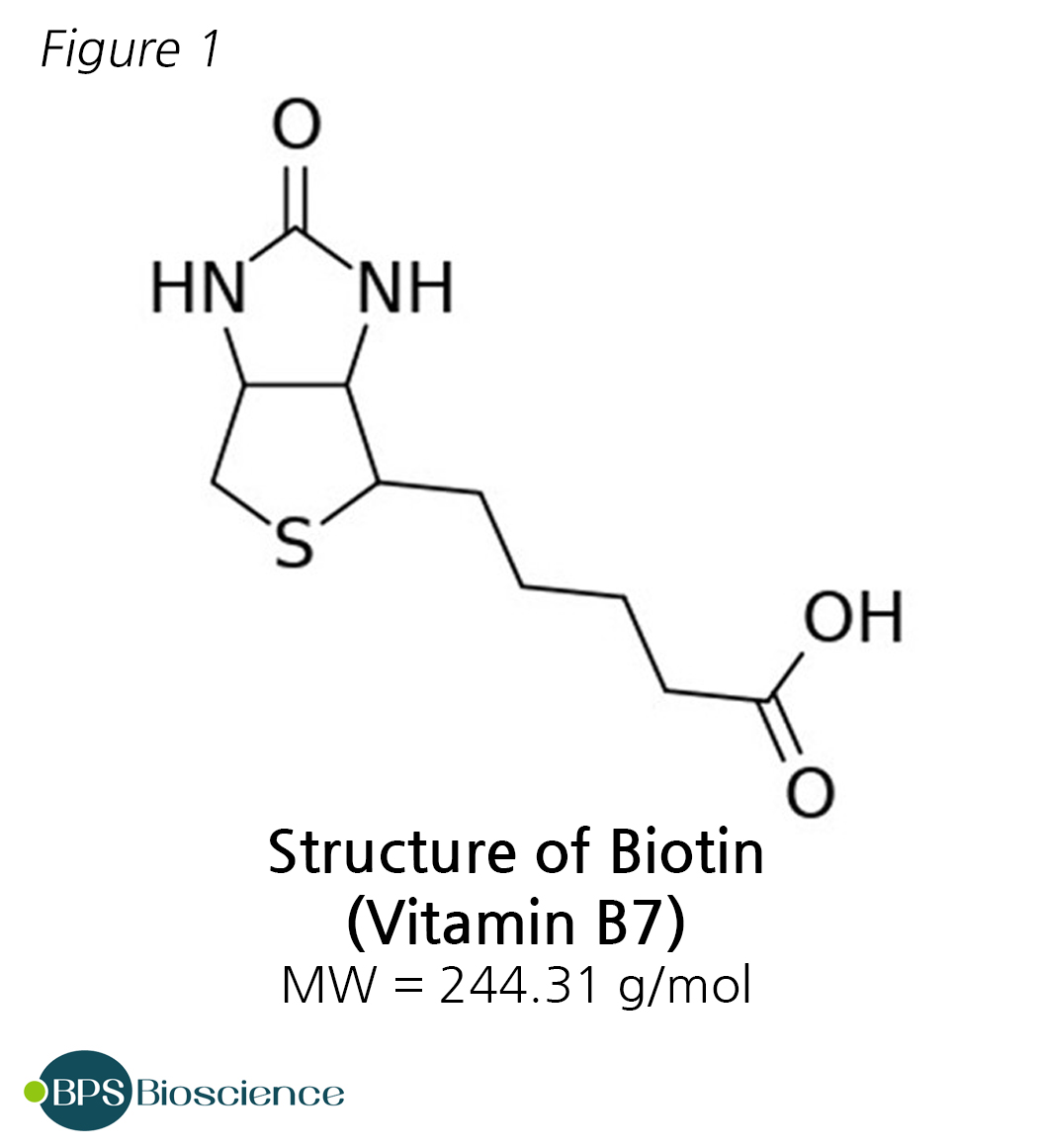 Biotin is a small, water soluble molecule of the vitamin B family. The most attractive property of biotin, from a biotechnological standpoint, is that it binds to avidin and to bacteria-derived streptavidin with an exceptionally high affinity and high specificity. Indeed, the binding of biotin and avidin is among the strongest noncovalent interaction known (dissociation constant in the range of 10−14 M to 10−15 M).
Biotin is a small, water soluble molecule of the vitamin B family. The most attractive property of biotin, from a biotechnological standpoint, is that it binds to avidin and to bacteria-derived streptavidin with an exceptionally high affinity and high specificity. Indeed, the binding of biotin and avidin is among the strongest noncovalent interaction known (dissociation constant in the range of 10−14 M to 10−15 M).
This extremely strong interaction is resistant to a variety of harsh experimental conditions (extremes of pH, temperature, buffer). Therefore, it is used extensively in scientific and biomedical applications to facilitate the detection, immunoprecipitation or purification of a molecule of interest. In addition, biotin has such a small molecular size that it is unlikely to perturb protein function dramatically.
Several molecules of biotin can be conjugated to a protein of interest. This is useful to amplify a weak signal, for example when very small amounts of protein are present, although it may come at the detriment of signal specificity since it leads to a high noise to signal ratio.
Biotinylation, the process of covalently attaching biotin to a molecule of interest, can be achieved either chemically or enzymatically. In contrast to chemical biotinylation methods, enzymatic biotinylation allows biotin to be linked at exactly one residue within the recognition sequence of a protein, with high uniformity, while allowing the pre-defined orientation of the subsequent avidin or streptavidin binding.
Enzymatic Biotinylation: BirA
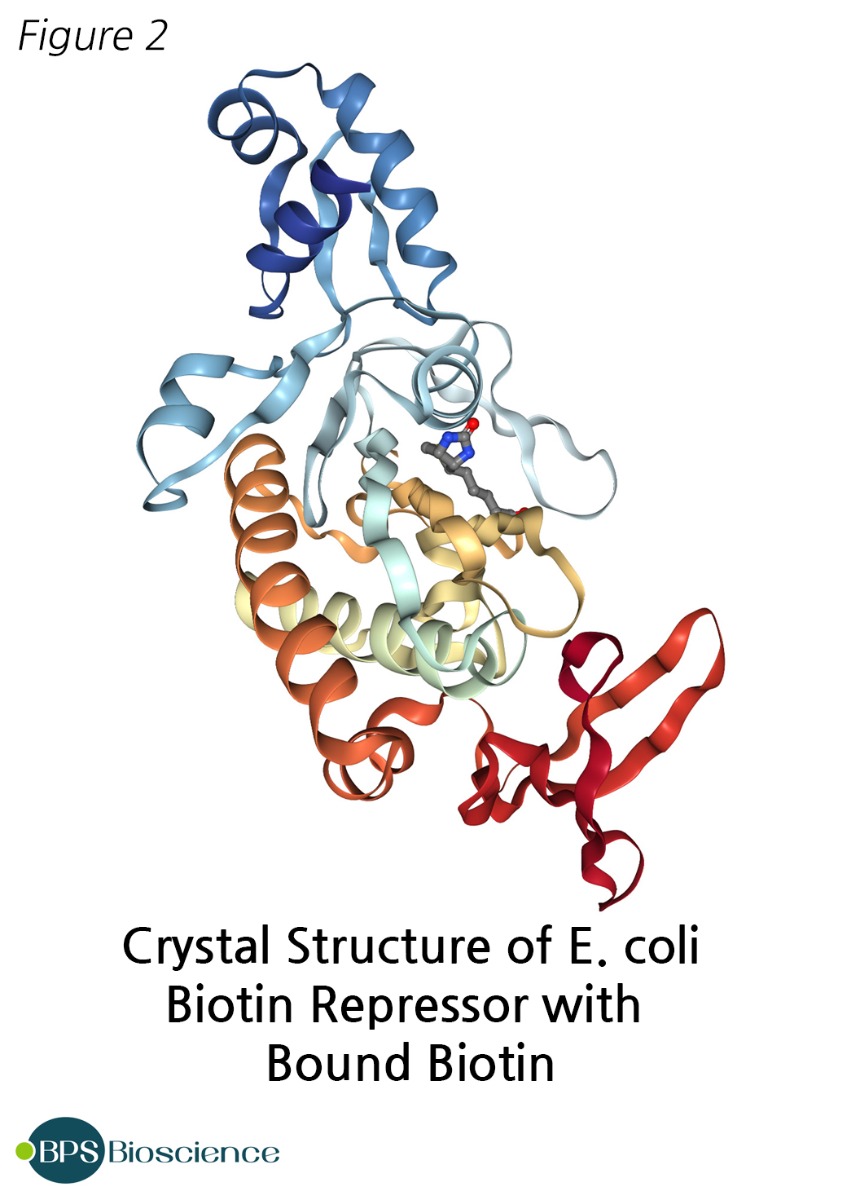 Ligation of biotin to a protein is a specific post-translational modification catalyzed by biotin ligase enzymes. The best characterized and most commonly used biotin ligase is protein BirA [P06709] from E. coli strain K12, which in bacteria also acts as the repressor of the biotin operon [1-3].
Ligation of biotin to a protein is a specific post-translational modification catalyzed by biotin ligase enzymes. The best characterized and most commonly used biotin ligase is protein BirA [P06709] from E. coli strain K12, which in bacteria also acts as the repressor of the biotin operon [1-3].
The publication of BirA full sequence in 1995 [NC_000913.3] resulted in its widespread use in biotechnology [4]. BirA, also known as biotin ligase, biotin operon repressor protein, biotin holoenzyme synthetase, and biotin-[acetyl-CoA carboxylase] synthetase, has a molecular weight of approximately 35kDa. Its 3-dimensional structure, determined at 2.3 Å resolution by X-ray crystallography, shows an asymmetric protein with three distinct domains [5, 6]. Additionally, it contains a GRGRRG motif associated with ATP binding, occurring in close proximity to the biotin binding site.
BirA catalyzes an ATP-dependent two-step reaction in which biotin is used to form biotinyl-5'-adenylate and is transferred to a specific lysine residue of the accepting protein via an amide linker.
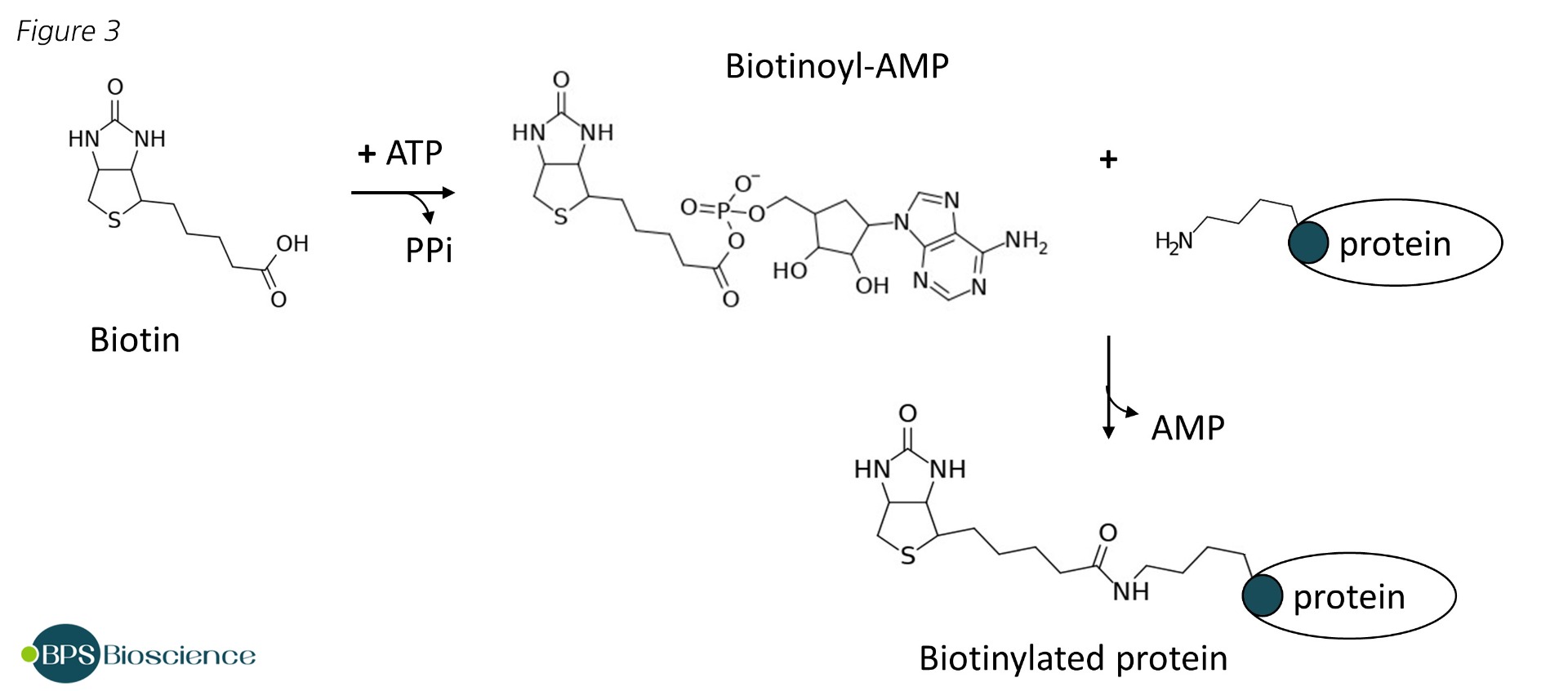
This is a reaction of stringent specificity that requires the recognition of a specific target sequence [7]. The natural substrate of BirA is the biotin carboxyl carrier protein (BCCP), which was formerly fused to proteins of interest before smaller tags were identified [8]. Screening of peptide libraries led to the first description of a consensus sequence of 13 amino acids sufficient for effective BirA mediated biotinylation [9]. Further optimization of this minimal peptide, notably with the addition of a glutamic acid in C-terminal, resulted in the final AviTag™ sequence GLNDIFEAQKIEWHE [10]. Kinetic experiments have demonstrated that BirA biotinylates the AviTag twice as fast as its natural substrate BCCP, and the AviTag™ is short enough to preclude significant alterations of function [11].
Today, fusing a protein of interest with an AviTag at a chosen location, usually but not necessarily N- or C-terminus, is the most common way of tagging proteins with biotin. BirA labels the lysine residue of the AviTag in a highly specific fashion in the presence of biotin and ATP, either in vitro or in vivo [12].
Conclusion
BirA is ideal to add biotin to a protein that contains an AviTag. The distinct advantage of enzymatic biotinylation using the AviTag is the control conferred on the location of the tag and the number of biotin molecules that can be appended. In this respect the enzymatic reaction is superior to chemical biotinylation using NHS-sulfo-biotin, which labels free amine groups randomly in the target protein and has the potential to disrupt the activity or the protein of interest.
Related Products and Services
To assist fellow scientists with biotinylation, BPS Bioscience has developed an in vivo biotinylation kit (BPS Bioscience #27461) containing all the components necessary to label biotin-acceptor molecules in chemically competent E. coli strain BL21. The bacteria contain an IPTG-inducible plasmid encoding ligase BirA and a resistance gene for streptomycin or spectinomycin. They can be transformed with a plasmid encoding the tagged protein of interest. Addition of IPTG induces the co-expression of BirA with the protein and results in biotinylation. Alternatively, the competent BL21 E. coli containing the IPTG-inducible BirA expression plasmid can be purchased on their own (BPS Bioscience #27462).
In vitro biotinylation of AviTag-fused proteins can be accomplished using purified recombinant BirA proteins, which have been tagged with GST, 6xHis and/or FLAG for convenient detection or for elimination of BirA by immunoprecipitation or affinity column (BPS Bioscience #70031, #70030, #70032).
Finally, BPS offers off-the-shelf biotinylated recombinants proteins, or will biotinylate proteins as part of our custom expression services.
References
- Barker DF, Campbell AM. The birA gene of Escherichia coli encodes a biotin holoenzyme synthetase. J Mol Biol. (1981) 146: 451-67. PMID: 7024555.
- Barker DF & Campbell AM. Genetic and biochemical characterization of the birA gene and its product: evidence for a direct role of biotin holoenzyme synthetase in repression of the biotin operon in Escherichia coli. J. Mol. Biol. (1981) 146: 468–492. PMID: 6456358.
- Eisenberg MA, Prakash O, Hsiung SC. Purification and properties of the biotin repressor. A bifunctional protein. J Biol Chem. (1982) 257: 15167-15173. PMID: 6129246.
- Howard PK, Shaw J, Otsuka AJ. Nucleotide sequence of the birA gene encoding the biotin operon repressor and biotin holoenzyme synthetase functions of Escherichia coli. Gene (1985) 35: 321-331. PMID: 3899863.
- Wilson KP, Shewchuk LM, Brennan RG, Otsuka AJ, Matthews BW. Escherichia coli biotin holoenzyme synthetase/bio repressor crystal structure delineates the biotin- and DNA-binding domains. Proc Natl Acad Sci U S A. (1992) 89: 9257-9261. PMCID: PMC50105.
- Weaver LH, Kwon K, Beckett D, Matthews BW. Corepressor-induced organization and assembly of the biotin repressor: a model for allosteric activation of a transcriptional regulator. Proc Natl Acad Sci U S A. (2001) 98: 6045-50. PMCID: PMC33419.
- Reed KE, Cronan JE Jr. Escherichia coli exports previously folded and biotinated protein domains. J Biol Chem. (1991) 266: 11425-11428. PMID: 2050659.
- Cronan JE Jr. Biotination of proteins in vivo. A post-translational modification to label, purify, and study proteins. J Biol Chem. (1990) 265: 10327-10333. PMID: 2113052.
- Schatz PJ. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: a 13-residue consensus peptide specifies biotinylation in Escherichia coli. Biotechnology (N Y). (1993) 11: 1138-1143. PMID: 7764094.
- Beckett D, Kovaleva E, Schatz PJ. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. (1999) 8: 921-929. PMCID: PMC2144313.
- Fairhead M, Howarth M. Site-specific biotinylation of purified proteins using BirA. Methods Mol Biol. (2015) 1266: 171-184. PMCID: PMC4304673.
- Cull MG, Schatz PJ. Biotinylation of proteins in vivo and in vitro using small peptide tags. Methods Enzymol. (2000) 326: 430-440. PMID: 11036656.







