Ubiquitination and CBL
Introduction
Ubiquitination is a process that involves the covalent binding of a 76 amino acid protein called ubiquitin to a target protein. This process is crucial for cellular homeostasis, regulating many cellular processes such as protein degradation, DNA repair, and cell signaling. Ubiquitination is a multistep process mediated by a group of enzymes known as ubiquitin ligases. Gain or loss of function of any of these enzymes can lead to impaired cellular functions and disease, such as cancer and immune disorders. On the other hand, controlled modulation of ubiquitination-regulating enzymes offers therapeutic opportunities.
Ubiquitination Mechanism
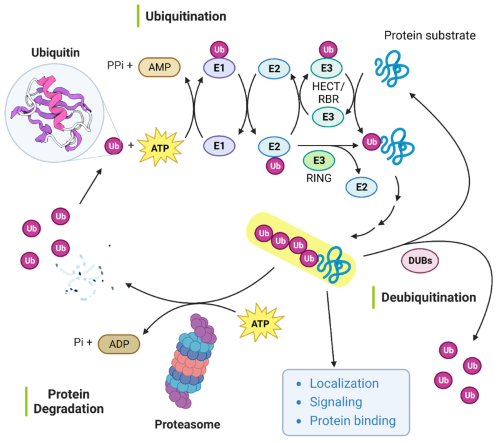
Figure 1: Protein ubiquitination and deubiquitination mechanism.
The first step in the process of ubiquitination involves the binding of ubiquitin (Ub) to an E1 Ub-activating enzyme. This step requires ATP hydrolysis to activate ubiquitin and forms a thioester bond between the E1 enzyme and ubiquitin. In the second step, ubiquitin is transferred to a ubiquitin-conjugating enzyme (E2), where Ub is also linked via a thioester bond. Finally, Ub is transferred from the Ub-E2 complex to a lysine residue in a substrate by two possible mechanisms. The transfer can occur directly from the E2 enzyme, in a reaction mediated by a RING E3 Ub ligase, or indirectly via Ub binding to a HECT/RBR E3 Ub ligase followed by transfer to the substrate (1). Ubiquitin can be added to proteins as a single ubiquitin monomer, or it can be added as chains of different lengths and linkage types, targeting proteins to different cellular fates. For example, poly-ubiquitinated proteins are typically targeted for proteosomal degradation or DNA repair functions, while mono-ubiquitation typically results in endocytosis (2, 3).
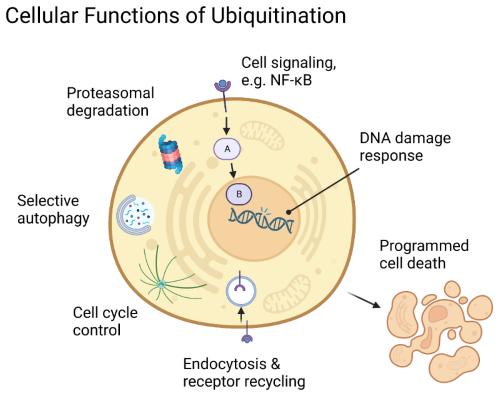
Figure 2: Cellular functions regulated by ubiquitination.
CBL
CBL proteins are RING finger E3 ubiquitin ligases. RING finger E3 ligases can function both as E3 ligases but also as adaptors, regulating a multitude of pathways. The CBL protein family is comprised of three members: CBL, CBL-b and CBL-c. They consist of an N-terminal tyrosine kinase binding domain (TKB domain), a RING finger domain, a proline rich region and a C-terminal ubiquitin-associated domain (UBA). The proline-rich region is involved in CBL’s role as adaptor. CBL and CBL-b are highly homologous and ubiquitously expressed, while CBL-c is a shorter form present primarily in epithelial cells (lacks the UBA domain). CBL’s main known function is the negative regulation of receptor and non-receptor tyrosine kinases (RTK and non-RTK, respectively), leading to their ubiquitination and degradation. CBLs exist in the cytosol as inactive proteins and are recruited to the tyrosine kinases (TK) upon stimulation by growth factors or cytokines. Cbl phosphorylation by the TK induces a conformational change that exposes the RING domain and allows E2 binding and TK ubiquitination. The Ub-TK complex is then retrieved from the plasma membrane and targeted for lysosomal degradation, leading to a negative regulation of TK-linked signaling pathways (4).
In addition to its direct role in the ubiquitin pathway, CBL can also function as an adaptor protein by binding Src homology region 2 and 3 (SH2 and SH3) domains in cell signaling proteins, permitting signal transduction (4).
Ubiquitination and CBL in disease and potential therapeutics
Given the importance of ubiquitination, it is clear that its dysregulation can lead to severe pathologies, as it results in abnormal protein misfolding, aggregation, and accumulation. For example, the accumulation of ubiquitin-conjugated protein aggregates is a pathological feature of Alzheimer's, Parkinson's, and Huntington's diseases. CBL malfunction can result in abnormal activation of RTKs or impact CBL adaptor activity. CBL, through negative regulation of receptors in T, NK and B cells, plays a role in innate and adaptive immune responses and is thus linked to cancer progression (3, 4, 5).
The use of anti-CTLA-4 and PD-1 in cancer immunotherapy has been widely studied, and immune checkpoints studies have now expanded into several other co-stimulatory and co-inhibitory molecular signals. However, limited effort has been put into studying modulating systems that orchestrate all those signals intracellularly, namely the ubiquitin pathway and CBL. In T cells, when CD28 is activated, CBL-b is ubiquitinated and targeted for lysosomal degradation, while in NK cells CBL-b regulates TAM (Tyro3, Axl and MER) receptor ubiquitination and internalization. Concomitant with CBL roles in immune cells, CBL-b knock-out mice showed an increased tumor rejection to implanted metastatic and non-metastatic tumors via activation of CD8+ T and NK cells (2). The targeting of CBL by small molecule inhibitors in high throughput screening identified highly selective inhibitors that increased IFNγ and degranulation of NK cells and T cell proliferation in a CD28-independent mode (6). Thus, inhibiting CBL-mediated protein degradation may be a useful strategy in improving immune function against cancers.
The use of PROTAC (Proteolysis-Targeting Chimera) for Targeted Protein Degradation (TPD) represents another opportunity for the use of E3 ligases for therapeutic use in diseases that involve proteins difficult to tackle by other approaches (7). PROTACs are composed of a ligand region that binds to the protein of interest, a linker, and then an E3 ligase and lead to target ubiquitination and subsequent degradation. The use of CBL in the context of a PROTAC may prove advantageous.
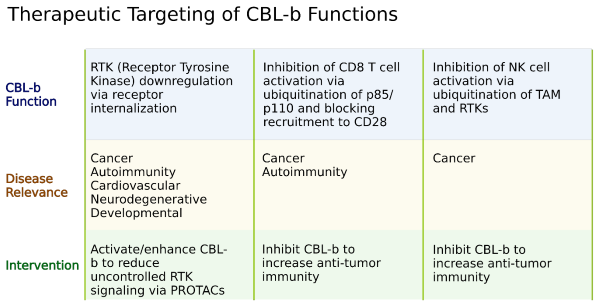
In conclusion, CBL and ubiquitin pathways are critical for cellular homeostasis, and dysfunction leads to a myriad of pathologies. Due to their roles, they have become promising therapeutical targets in cancer immunotherapy, either alone or in combination with other existing therapeutical approaches.
References
(1) Damgaard, R., 2021 Cell Death & Differentiation. 28: 423-426
(2) Tang R., et al., 2019 Cell Immunol. 340: 103878
(3) Liu J., et al., 2021 Signal Transduction and Targeted Therapy. 6: 28
(4) Leardine D., et al., 2022 Cancers (Basel). 14 (3):839
(5) Guo X., et al., 2021 J Immunother Cancer. 9 (3): e001975
(6) Riling C., et al., 2018 Mol Cancer Ther. 17: A206
(7) Bekes M., et al., 2022 Nature Reviews Drug Discovery. 21:181-200
│Related Products
- Assay Kits
- Inhibitors
- Activating E1 Enzymes
- Conjugating E2 Enzymes
- E3 Ligases
- Deubiquitination Enzymes

