SARS-CoV-2 Coronavirus & COVID-19
There are seven known human coronaviruses, including the novel SARS-CoV-2. Four of these strains cause only mild respiratory symptoms; however, coronaviruses became notorious after outbreaks of Severe Acute Respiratory Syndrome (SARS) in 2002 and Middle East Respiratory Syndrome (MERS) in 2012. The SARS-CoV-2 (2019-nCoV) coronavirus, first identified in late 2019, causes severe pneumonia and respiratory infection (COVID-19).
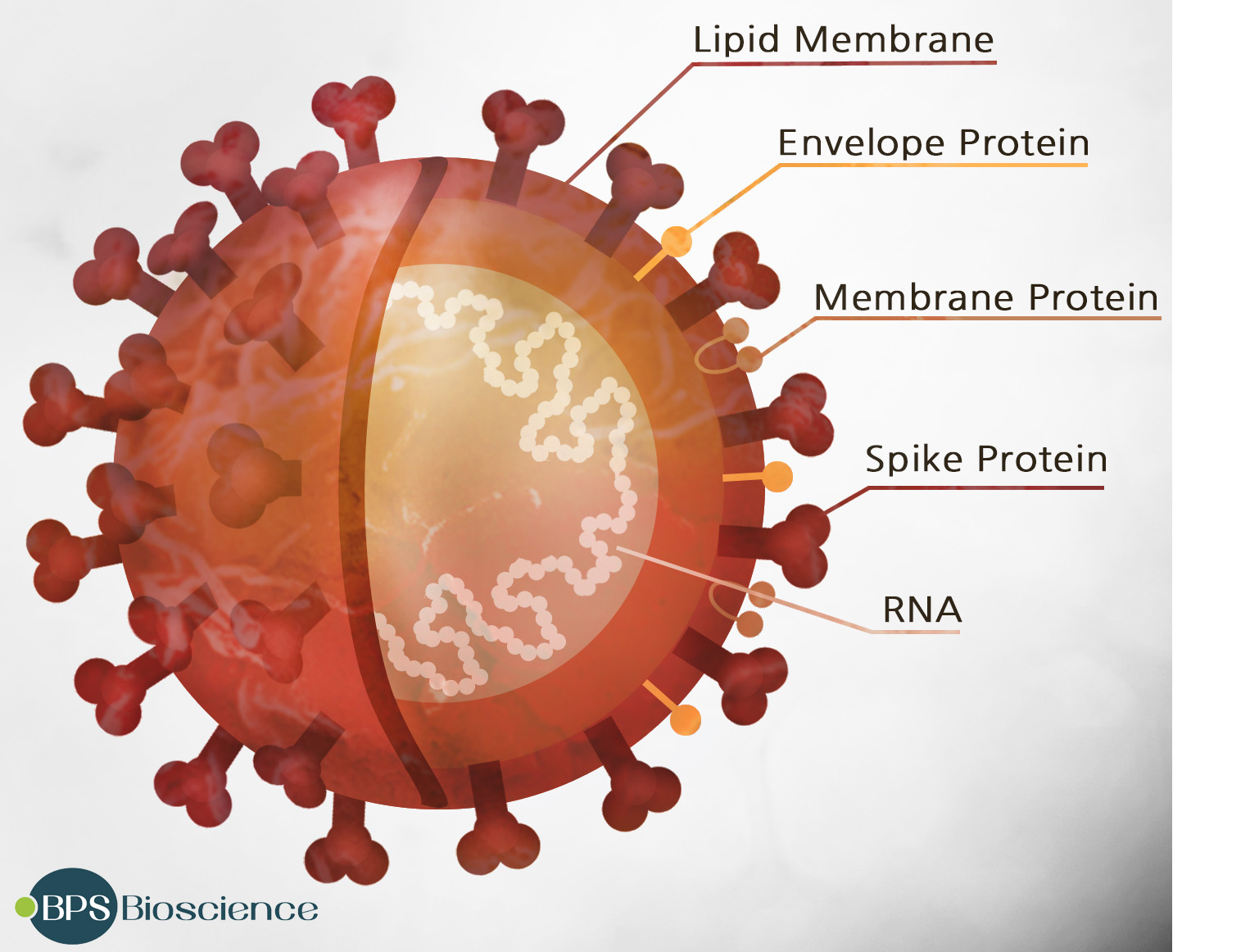
The SARS-CoV-2 virus consists of four different proteins and a strand of RNA. The most prominent feature, a trimer formed by the “Spike” protein, sticks out from the membrane and gives the virus its distinctive “corona” structure. Two other proteins, envelope protein and membrane protein, reside in the membrane between these spikes, providing structural integrity. Inside the membrane, a fourth protein, nucleocapsid, acts as a scaffold surrounding the 29,900 nucleotides of RNA making up the viral genome.
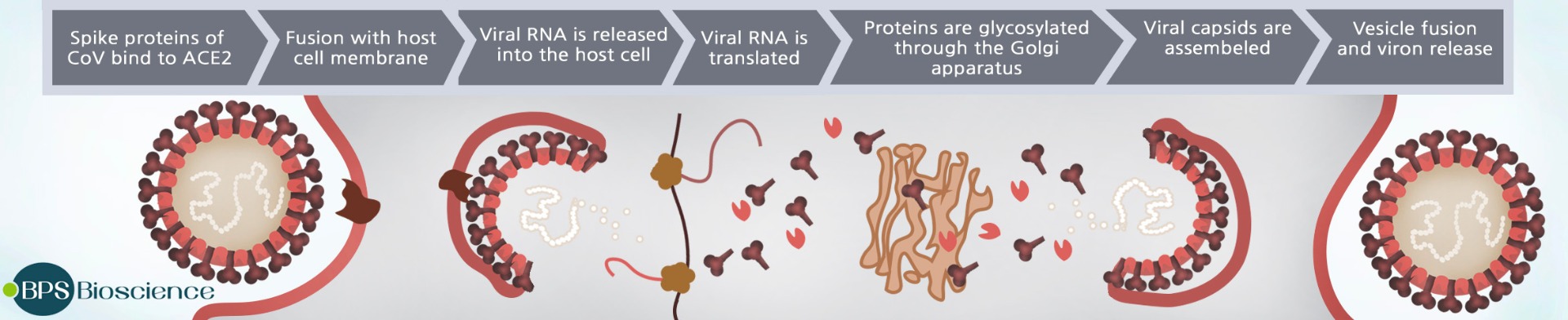
SARS-CoV-2 enters target cells by binding the coronavirus Spike protein to ACE2 (angiotensin converting enzyme 2), a homodimer found on the surface of some human cells, particularly those in the respiratory tract. (2) The Spike protein consists of two subunits, S1 and S2, and the S1 subunit has a receptor binding domain (RBD) which interacts with ACE2 to facilitate viral attachment. Modeling based on the crystal structure suggests simultaneous binding of two Spike protein trimers to the ACE2 dimer. (3) Following this attachment, the cellular proteases furin and TMPRSS2 cleave the Spike protein, and the truncated S2 subunit of the Spike protein facilitates fusion of viral and cellular membranes, bringing the virion into the cell. (4) Both ACE2 and TMPRSS2 are coexpressed by type II pneumocytes, which represent important viral target cells. Once the virus enters the cell, the viral RNA is translated to encode the viral replicase gene and two large polyproteins that are processed by viral proteases (Papain-like protease 2 (PL2pro) and 3C-like protease (3CLpro, also referred to as main protease, Mpro). These cysteine proteases cleave the polyproteins into 15–16 nonstructural proteins involved in viral RNA synthesis and replication. (5) The efficiency of Spike binding to ACE2 was found to be a key to the transmissibility of SARS-CoV, the related coronavirus responsible for the global spread of SARS in 2003 (6). Recent studies show the novel SARS-CoV-2 binds to ACE2 on human cells with at least 10x higher affinity than SARS-CoV. (7) This difference in affinity may help explain why COVID-19 infection is so much more contagious than SARS.
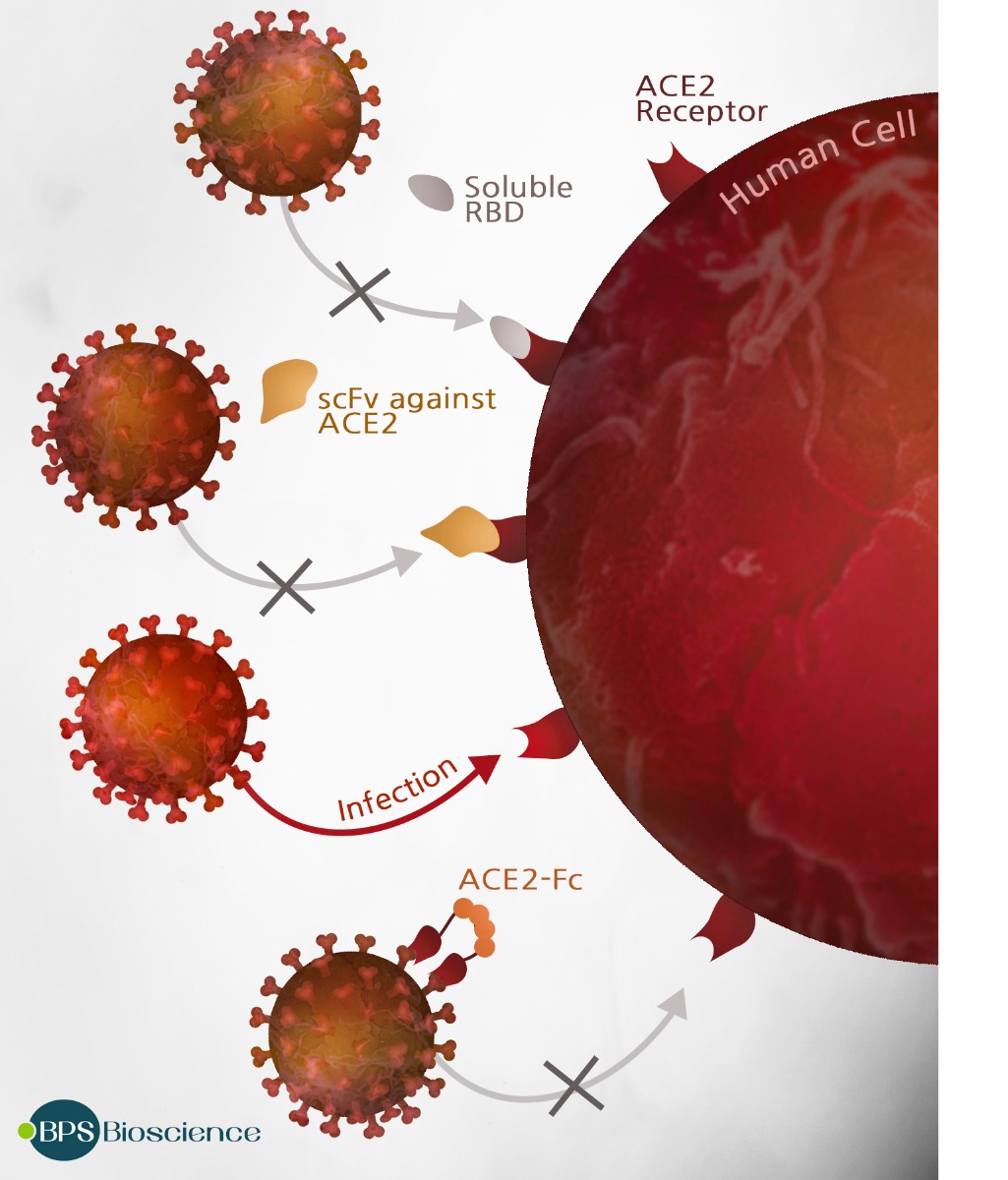
Researchers are actively working to develop a vaccine and antiviral drugs that can block coronavirus infection or replication. One interesting line of attack involves using ACE2 fragments as a decoy to competitively prevent the coronavirus from binding to lung cells. (8) Alternately, fragments of the Spike protein corresponding to the receptor binding domain (RBD) might be used as a peptidomimetic antagonist to block ACE2 sites on cells and prevent viral attachment. Other approaches target the Spike protein directly to prevent its interaction with ACE2. Passive immunization of COVID-19 patients with convalescent-phase sera suggests that immunotherapeutic approaches using neutralizing monoclonal antibodies that bind the Spike RBD have significant therapeutic or vaccine potential.
Immunomodulatory therapy may also provide insights into the treatment of COVID-19. In many COVID-19 infected patients, the overproduction of early response proinflammatory cytokines (cytokine storm) leads to rapid, severe pathogenesis and high mortality. Immunomodulatory agents that directly target the key cytokines involved in cytokine storm may help to alleviate this hyperinflammation. IL-6 can induce severe inflammation in the lungs of patients with COVID-19, and elevated levels of IL-6 in the blood have been reported to predict a fatal outcome. (9) Tocilizumab (by Roche/Chugai), a recombinant monoclonal antibody that blocks IL-6 receptor, has been used successfully to treat cytokine release syndrome in CAR T-cell immunotherapy, and is currently under evaluation for its ability to block inflammation in COVID-19 infections.
Proteolytic cleavage of the Spike protein into the S1 and S2 domains is critical for viral fusion into the cellular membrane, making the cellular proteases furin and TMPRSS2 interesting therapeutic targets. (10) Since these two serine proteases cleave different locations on the Spike protein, small molecule protease inhibitors such as camostat mesylate (TMPRSS2) and MI-1851 (furin) can work synergistically to prevent viral entry. Another promising drug target is the 3CL protease, which processes the polyproteins that are translated from the viral RNA. 3CL protease is critical for viral replication, making it an attractive target for new anti-coronavirus drugs. An additional advantage of this approach is that there are no human proteases with similar cleavage specificity, so any inhibitors of 3CL are unlikely to be toxic. (11)
While the highly touted hydroxychloroquine showed unimpressive results in clinical trials, positive results were seen with the investigational compound Remdesivir (by Gilead). Remdesivir is a nucleotide analog that incorporates into RNA and disables RNA-dependent RNA polymerases. Originally designed as an Ebola drug, Remdesivir had been used successfully to inhibit the replication of the related coronaviruses SARS and MERS in tissue culture and animal models.(12) Preliminary results of trials with patients suffering from COVID-19 indicate that patients receiving Remdesivir have 31% faster recovery time than those who receiving a placebo (p<0.001). (13) A similar RNA polymerase inhibitor called Favipiravir (by Toyama), originally developed as an anti-influenza drug, is also undergoing Phase III clinical trials as a treatment for COVID-19.
The landscape for coronavirus diagnostics, vaccines, and treatments is changing daily, as scientists work closely with medical professionals to battle the SARS-CoV2 virus. Researchers have made rapid progress in the characterization of the novel coronavirus and are continuing to work swiftly and diligently to develop innovative therapies and vaccines to defeat the virus. The urgency of the current pandemic requires no less.
BPS Bioscience continues to be a leader in providing products and services to help meet the urgent need for new drugs and vaccines to treat and prevent COVID-19. BPS Bioscience has quickly developed an entire portfolio of recombinant proteins, unique assay kits, pseudovirions, antibodies, and lentiviruses to help researchers understand the pathogenesis of viral disease and to advance research and development of therapeutic drugs and vaccines. Visit our website frequently to discover our newest product and service offerings.
References:
- https://www.worldometers.info/coronavirus
- Ou, X., et al. 2020. Nat Commun 11: 1620. doi:10.1038/s41467-020-15562-9
- Yan, R., et al. BioRxiv. 2020 Feb 18; in press. doi:10.1101/2020.02.17.951848
- Hoffmann et al., 2020, Cell 181(2):271-280.e8. doi: 10.1016/j.cell.2020.02.052
- Zhang, L., et al. Science 20 Mar 2020:eabb3405 in press. doi: 10.1126/science.abb3405
- Li, W. et al., 2005. EMBO J. 24:1634-1643
- Wrapp, D., et al. 2020. Science 367(6483): 1260-1263
- Renzi, F and Ghersi, D. BioRxiv. 2020. Preprint doi: 10.1101/2020.04.06.028647.
- Zhao, M. Int J Antimicrob Agents. 2020 Apr 16:105982. doi:10.1016/j.ijantimicag.2020.105982
- Bestle, D., et al. BioRxiv. 2020. Preprint. doi: 10.1101/2020.04.15.042085
- Zhang, L. et al. 2020. Science 368(6489): 409-412.
- Gordon, CJ., et al. 2020. J. Biol. Chem. 295: 4773-4779.
- https://www.niaid.nih.gov/news-events April 29, 2020.