ADAR1 Activity Luciferase Reporter HEK293 Cell Line
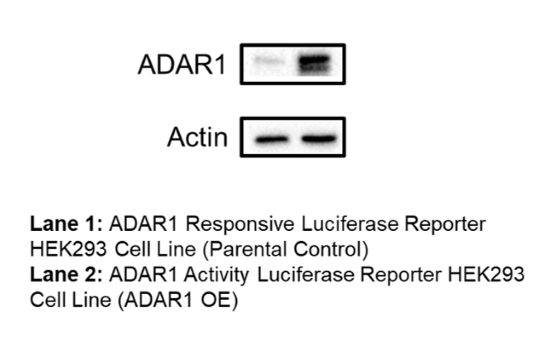
ADAR1 Activity Luciferase Reporter HEK293 Cell Line is a HEK293 cell line designed to monitor the activity of ADAR1 (adenosine deaminase acting on RNA) enzyme. These cells were engineered to express ADAR1 (NM_001111.5) and an ADAR1 reporter construct comprised of an ADAR1 hairpin target with a stop codon (UAG) susceptible to ADAR1-mediated editing to tryptophan (UUG), located upstream of a firefly luciferase reporter (Figure 1).
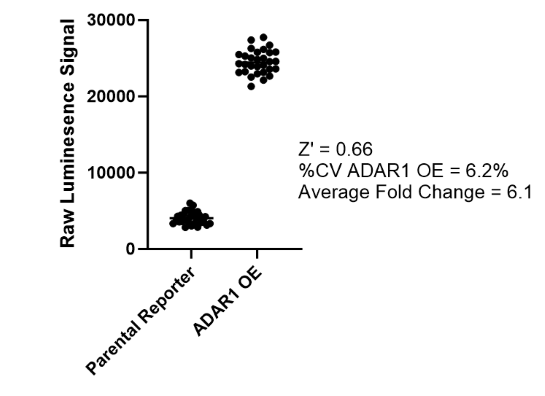
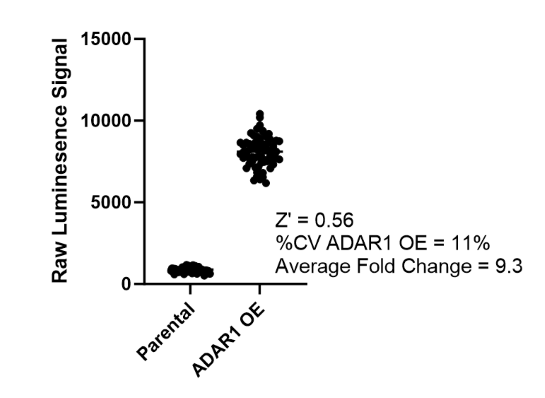
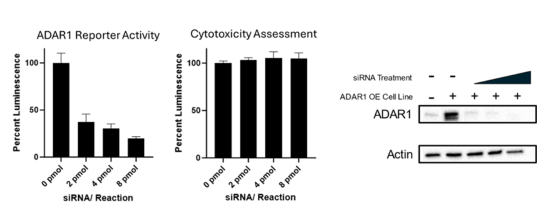
This cell line has been validated by comparing reporter activation to ADAR1 Responsive Luciferase Reporter HEK293 Cell Line (#82238) and in response to treatment with ADAR1 siRNA.
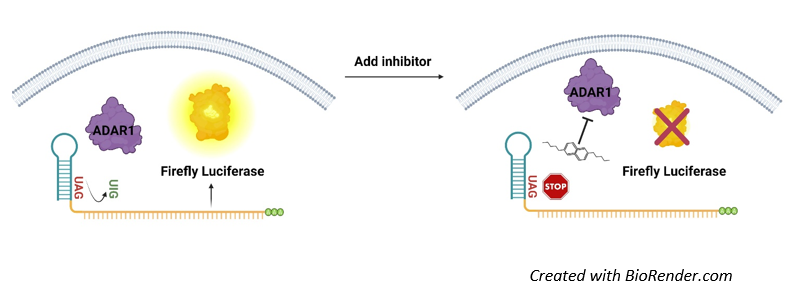
Figure 1: Illustration of the mechanism of action of ADAR1 Activity Luciferase Reporter HEK293 Cell Line.
The ADAR1 reporter construct is comprised of an ADAR1 hairpin target with a stop codon (UAG) upstream of the sequence encoding luciferase. In the absence of ADAR1, luciferase is not transcribed, and the cells show no luciferase activity. In the presence of ADAR1 activity, adenine is converted into inosine, encoding now the amino acid tryptophan (UUG) and enabling transcription and expression of luciferase. ADAR1 activity, therefore, directly correlates with luciferase activity.
Interested in screening and profiling inhibitors or activators of ADAR1 without the need to purchase and license the cell line? Check out our Cell Signaling Pathway Screening and Profiling Service.
Purchase of this cell line is for research purposes only; commercial use requires a separate license. View the full terms and conditions.
Materials Required for Cell Culture
| Name | Ordering Information |
| Thaw Medium 1 | BPS Bioscience #60187 |
| Growth Medium 1U | BPS Bioscience #78548 |
Materials Required for Cellular Assays
| Name | Ordering Information |
| ADAR1 Responsive Luciferase Reporter HEK293 Cell Line | BPS Bioscience #82238 |
| ADAR1 Targeting siRNA | Horizon #M-008630-01-0005 |
| Lipofectamine™ RNAiMAX Transfection Reagent | Thermo Fisher #13778030 |
| Assay Medium 1A | BPS Bioscience #79805 |
| Thaw Medium 1 | BPS Bioscience #60187 |
| 96-well tissue culture white, clear-bottom assay plate | Corning #3610 |
| ONE-Step™ Luciferase Assay System | BPS Bioscience #60690 |
| Luminometer |
The cell line has been screened to confirm the absence of Mycoplasma species.
ADAR (Adenosine Deaminase Acting on RNA) enzymes perform adenosine to inosine base editing in RNA, particularly targeting adenosines located within a specific double-stranded stem-loop motif (Figure 1). In the context of healthy, uninfected cells, ADAR1 performs A-to-I editing on endogenous double-stranded RNA to prevent it from activating the downstream dsRNA sensors RIG-I (retinoic acid-inducible gene I) and MDA5 (melanoma differentiation-associated protein 5), which in-turn activate a pro-inflammatory response. Loss of function mutations in ADAR1 result in aberrant activation of the dsRNA sensors and are involved in autoimmune disorders. ADAR1 dysfunction also impacts cancer cell growth, proliferation, and response to immunotherapy. ADAR1 expression is increased in many tumor types and ADAR1 knock-out has shown to improve the response to certain immunotherapies such PD-1 (programmed death protein 1)/PD-L1 (programmed death ligand 1) blockade and to circumvent tumor immunotherapy resistance mechanisms, making ADAR1 an attractive target for therapeutic development.
Bhate A., et al., 2019 Mol Cell. 73(5):866-868.
Buchumenski I., et al., 2019 Nature. 565(7737):43-48.
Gallo, A., et al., 2017 Human Genetics 136(9): 1265–1278.
Ishizuka JJ, Manguso RT, Cheruiyot CK, Bi K, Panda A, Iracheta-Vellve A, Miller BC, Du PP, Yates KB, Dubrot J,
Xu L. and Öhman M., 2018 Genes (Basel). 10(1):12.
Yuan, J., et al., 2023 J Exp Clin Cancer Res 42:149.
Zhang, T., et al, 2022 Nature 606: 594–602.