Posters
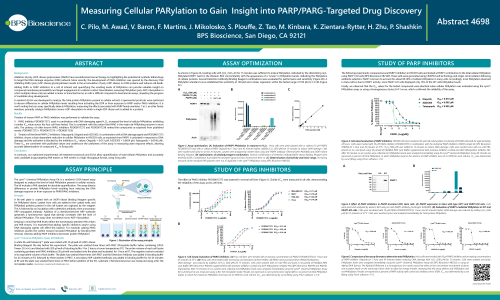
Inhibitors of poly (ADP-ribose) polymerases (PARPs) have revolutionized cancer therapy by highlighting the potential of synthetic lethal drugs to target the DNA damage response (DDR) network. More recently, the development of PARG inhibitors was spurred by the discovery that inhibiting PARG (poly (ADP-ribose) glycohydrolase) results in the accumulation of poly (ADP-ribose) on DDR proteins and induces cell death.
Adding PARG or PARP inhibitors to a cell of interest and quantifying the resulting levels of PARylation can provide valuable insight on compound membrane permeability and target engagement in a cellular context. Nevertheless, assessing PARylation in cells is difficult, impeding the progress of candidate drug development. A sandwich ELISA was developed to analyze the total protein PARylation present in cellular extracts. Experimental protocols were optimized to discern differences in cellular PARylation levels resulting from activating the DDR or from exposure to PARP and/or PARG inhibitors.
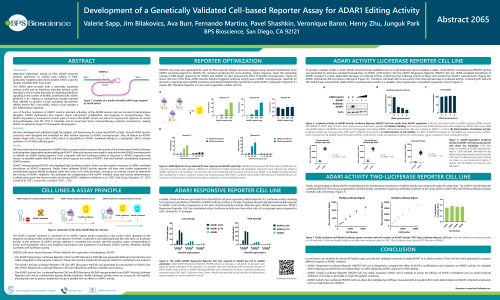
Adenosine Deaminase Acting on RNA (ADAR) enzymes perform adenosine to inosine base editing in RNA, particularly targeting adenosines located within a specific double-stranded stem-loop motif. ADAR editors are promising therapeutic targets and are used as tools in experiment treatments for genetic diseases. We describe the design and optimization of three cell lines generated to support different aspects of ADAR1 research:
ADAR1 Responsive Luciferase Reporter Cell Line is designed to compare the effect of ADAR1 modifications and mutations on ADAR1 activity, for example when studying structure/function relationships, or when designing ADAR1 variants for RNA editing.
ADAR1 Activity Luciferase Reporter Cell Line stably expresses ADAR1 and is suitable to assess the efficacy of ADAR1 modulators such as small molecule inhibitors. This assay is amenable to high-throughput screening.
ADAR1 Activity Two-Luciferase Cell Line allows the multiplexing of efficacy measurements in parallel with toxicity determination of ADAR1-directed compounds such as small molecule inhibitors.

Better control of blood glucose levels in diabetic patients has been achieved with GLP-1R (glucagon-like peptide-1 receptor) agonists, which also promote a sense of satiety, helping patients with weight loss and reducing cardiovascular risks. Dual or triple receptor agonists targeting GLP-1R, GCGR (glucagon receptor) and GIPR (glucose-dependent insulinotropic polypeptide receptor) may provide even better options to improve metabolism and health outcomes.
To support multi-receptor agonist development, we generated and characterized three cell lines that respond to GCGR, GIPR, or GLP-1R activation by inducing luciferase activity which can be measured using a simple luminometer and is directly proportional to receptor activation. These cells express GCGR, GIPR, or GLP-1R, and a firefly luciferase reporter under the control of CRE (cAMP response elements). They produce a robust luciferase readout upon agonist stimulation (up to 40 to 300-fold stimulation depending on the cell line) and are ideal for screening candidate molecules and determining their EC50.

Proprotein convertase subtilisin/kexin type 9 (PCSK9) regulates circulating cholesterol levels by binding to low-density lipoprotein receptor (LDLR) on the cell surface. Gain-of-function (GOF) mutations in PCSK9 increase the risk of cardiovascular disease, and PCSK9 has become a promising therapeutic target for the treatment of inherited or sporadic cardiovascular disease. Two assays were designed to evaluate the effect of candidate drugs on PCSK9/LDLR binding. Experimental parameters and buffer compositions were optimized to detect conformational effects of pH and PCSK9 mutations. Our experimental findings are consistent with existing literature on PCSK9 mutations, demonstrating the utility of these assays. Optimizing drug candidates to function effectively across a range of pH conditions enhances their potential clinical applicability. Thus, measuring PCSK9-LDLR binding at different pH levels is crucial for understanding the physiological relevance, optimizing drug development, unraveling intracellular trafficking mechanisms, exploring disease mechanisms, and gaining basic biological insights into this important molecular interaction.

Poly-(ADP-ribose) glycohydrolase (PARG) degrades ribosylation chains generated by PAR polymerases PARPs) in response to DNA damage and has been implicated in cancer. PARG is a promising therapeutic target as its inhibition may potentiate the effect of chemotherapy and radiation. This study describes the optimization of two biochemical assays measuring PARG enzymatic activity for inhibitor screening & profiling. The PARG Fluorogenic Assay uses a straightforward, fast, no-wash method to study enzyme kinetics, and is ideal for high-throughput screening applications. However, test compounds with intrinsic fluorescence will interfere with the assay, requiring a non-fluorescent method. The PARG Chemiluminescent Assay, although inappropriate for kinetic studies, offers a convenient alternative to the fluorogenic assay.

Casitas B-lineage lymphoma (CBL) is a family of RING-type E3 ligases that target proteins for degradation via the proteasomal pathway. CBL-b specifically functions as a negative effector of T cell activation by down regulating the T cell receptor, whereas c-CBL interacts with various receptor tyrosine kinases involved in cell signaling and activation, targeting them for degradation. In this study, we used TR-FRET to uncover new mechanistic aspects of CBL activation. Our data indicates that no significant difference in the parameters of substrate ubiquitination exists between CBL-b and c-CBL, and that ubiquitination of receptor tyrosine kinase Tyro3 by either c-CBL or CBL-b depends on the kinase binding to CBL but not on the phosphorylation of CBL.
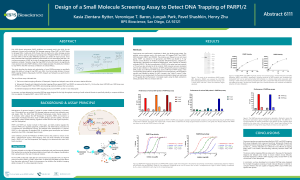
Poly (ADP-ribose) polymerase (PARP) inhibitors are currently used in the clinic for the treatment of tumors with a defective DNA damage response. The continuous presence of PARP at the site of damage prevents repair and blocks replication, leading to cell death. Therefore, drugs that trap PARP1/2 to the DNA tend to be significantly more cytotoxic than other PARP inhibitors, which is highly desirable. This study describes the design and optimization of novel PARPtrap assays to specifically assess the ability of a drug to trap PARP onto DNA.

Drug discovery and development projects require reliable assays to screen and evaluate new small molecules that potentially inhibit RAS isoforms. We describe the development of two types of assays for high-throughput screening applications and titration of candidate compounds. One assay is based on AlphaScreen® technology, while the other takes advantage of BODIPY®-GDP in fluorophore GDP/GTP exchange assay kits.

Designing new therapeutic strategies requires the generation of appropriate tools, which can use considerable time and resources. BPS Bioscience has generated a varied portfolio of validated immuno-oncology tools to support drug development efforts as well as basic research projects, allowing the scientific community to focus on critical questions. These tools include anti-BCMA antibodies for BCMA detection, BiTEs (Bispecific T cell Engagers) and TriTEs, BCMA cell lines with or without reporter genes, biochemical assays, and BCMA lentivirus. Thus, BPS supports researchers at all phases of drug discovery to accelerate the development of new treatments for human diseases.

PROTACs® (Proteolysis Targeting Chimeras) are heterobifunctional molecules that bind a target protein of interest (POI) and a specific E3 ligase, triggering the target for degradation through the ubiquitin-proteasome system. This therapeutic approach provides advantages over classical drug therapy enabling targeting of otherwise undruggable proteins, while requiring relatively low amounts of compound. To support the advancement of therapeutic PROTACs, we have developed biochemical assays using AlphaScreen™ technology, providing a simple, rapid, and sensitive no-wash system for high throughput-screening.

