The Impact of Cell Culture Conditions and Media in Cellular Studies
Cell Culture
The first attempt to keep cells alive outside of the body traces back to the 19th century, when Sydney Ringer created a buffer able to maintain cardiac cells beating in vitro. Cell culture allowed the study of cellular responses and mechanisms under defined and simplified conditions. While it is a standard technique today, one should not forget that it takes cells out of their biological context, creating an artificial system, and the responses and phenotypes observed may not fully represent the situation in the body. The CAR-T field of research, for example, has struggled with the translation of findings from in vitro to in vivo, due to the inability to mimic the tumor microenvironment in a 2D system. Researchers have been working on designing more complex systems that still allow studies outside of the human body but get closer to situations in vivo, such as organoids. Organoids are 3D cellular systems that recapitulate spatial organization of heterogeneous tissue-specific cells, cell- cell, cell-matrix interactions. Either in the context of an organoid or a 2D system, cells are exposed to cell culture conditions, such as cell culture media, and their characteristics can be altered significantly by variations in these conditions.
Cells are highly responsive to their environment, both in vivo and in vitro. During the cell culture process, cells are typically exposed to a certain pH, osmotic pressure, temperature, a matrix and media. In the case of primary cells, isolation methods can often also have an impact (1). Cell culture media are designed to provide essential nutrients such as amino acids, carbohydrates, vitamins, growth factors, hormones, etc. Complete media are usually formulated as basal media, which are then supplemented with a more complex source of essential molecules, often times undefined, such as FBS (fetal bovine serum).
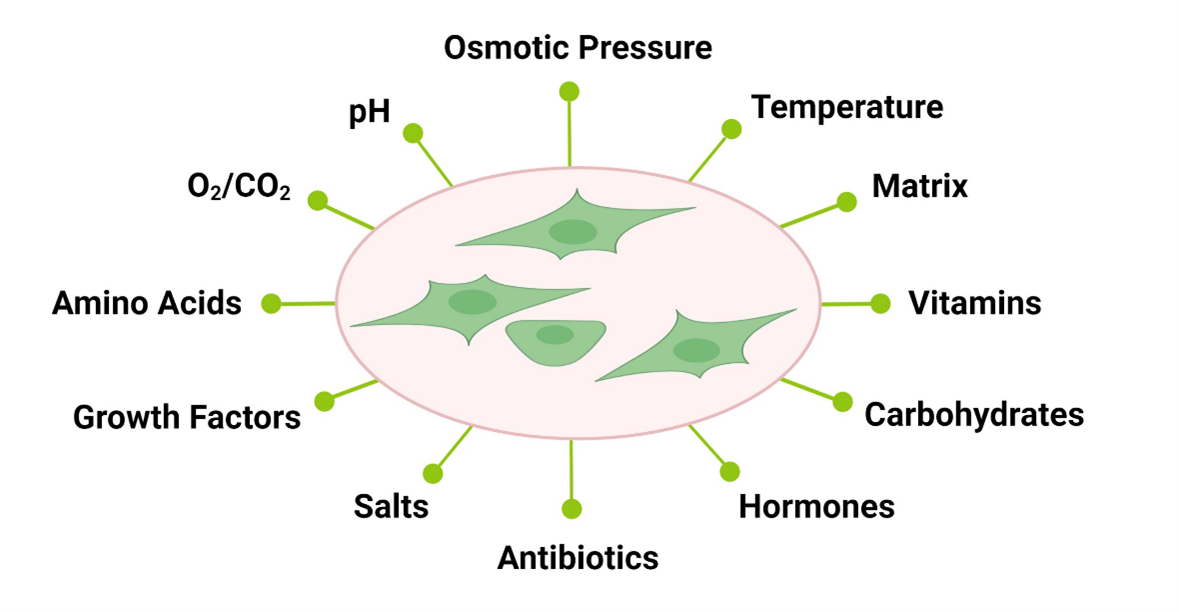
Figure 1: In vitro cell culture exposes cells to a broad range of environmental cues to which they respond.
Impact of Media in Cell Culture Outcomes
The results obtained with any cell culture system is dependent on three inter-related aspects: cell source, cell culture media, and culture conditions (example, static versus stirred systems). Only a synergy of these factors can result in the desired outcome, and changes in any will need adjustment of the others. Some of the impacts can be easily detected, as when cell morphology changes in response to culture conditions, but others require closer analysis (such as differential expression of cell markers and changes in metabolism) (1,2).
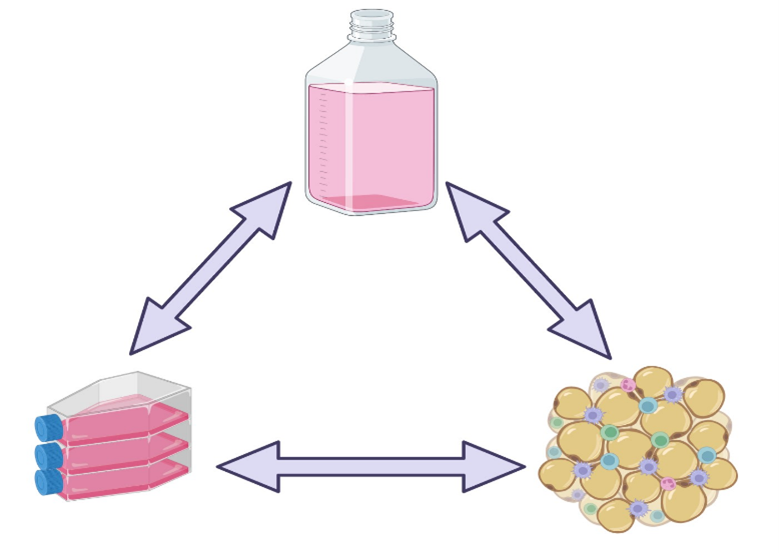
Figure 2: Experimental observations obtained from cell culture systems are the result of a close relationship between cell source, cell culture system and cell culture media.
Alterations in cell metabolism are often found in cancer and inflammatory diseases and are crucial to support the new energetic needs of these abnormal cells. Cancer drug discovery workflows start with cell culture models, and often those studies are the basis for the identification of lead molecules and assessment of potency and toxicity profiles. Researchers studying cellular metabolism often struggle with cell culture variability, largely coming from the variation of the metabolite composition of media. The impact of cell culture media can be as dramatic as to lead to different cancer drug sensitivities and performances, which have obvious consequences during drug development (2,3).
The impact of media is seen not only in primary cell cultures but also in stable cell lines, as observed for CFBE41o- cells (stable human airway cell line). Differences were seen in cell phenotype, CFTR (cystic fibrosis transmembrane conductance regulator) expression and function. These differences can have a severe impact in studies on cystic fibrosis (4).
Animal-Derived Serum, A Complex Mixture of Reagents
One component in particular is responsible for large variability in cell culture outcomes: animal serum. Fetal, newborn or calf bovine serum are animal-derived sera commonly used in cell culture. While one tends to look at it as a single component, animal serum is in truth made of hundreds of identified and non-identified components present in unknown amounts. It is a highly variable mix, as unique as the animals that were used in its production. The sex of the animal, diet, country, strain, and genetics are all contributing factors to serum composition. This diversity in formulation can in turn lead to different cell characteristics with each lot of serum, and to an inability to reproduce scientific findings between laboratories, or even in the same laboratory over time.
Attempts to create serum-free media, which could result in more controlled and reproducible cell culture systems, had moderate success depending on the cell type of interest. Since the composition of animal serum is not fully known and we are still in the mist of understanding all the nuances and players involved in molecular pathways, it is inevitable that some components of importance are excluded in the serum-free formulations. In addition, serum-free media tend to be quite specific in their application, removing the flexibility of using the same formulation for several cell types. Animal serum, with its broad composition, ends up accommodating to the needs of multiple cell types, while serum-free medium is stripped down to its basic components.
Examples of the impact of serum in cell culture can be found in the literature. Evaluation of several lots of FBS in the culture of ARPE-19 (adult retinal pigment epithelial) cells resulted in different confluency. This correlated with the presence of certain growth factors in one lot and absence from other lots (5). Another example with high clinical relevance relates to the impact of serum in the culture of MSCs (mesenchymal stem cells). MSCs are known to vary with each donor and tissue used as source, but it has been found that a potential player in the divergent clinical outcomes observed in the use of MSCs in the clinic is actually the FBS used during their culture. One study found that the type of serum used in the culture of MSCs created specific niche conditions, which translated into a stimulatory or inhibitory activity of MSC towards HSC (hematopoietic stem cell) generation, and identified 5 crucial upstream signals that were impacted (Tp53, TGFB1, RABL6, ATF4 and TRIB3) and 785 genes with significant different patterns of expression. These major differences can have a severe impact on the ability of HSC to repopulate the bone marrow in animal models (6). Discrepancies in outcomes between studies can invalidate a therapy that may have tremendous patient benefit, and end-up reducing treatment options. The choice of serum is thus not a trivial aspect and should be considered with extreme care.
The question then becomes: how can one minimize the impact of using an undefined and variable reagent and still take advantage of its beneficial properties? It is unavoidable that variation will occur, however, performing animal serum lot validation and acquiring a complete lot can support your research for a long time. This is a costly, time-consuming process that requires a comprehensive testing of the cellular responses of interest to the different lots of serum, and the investment to purchase and store a complete lot of serum. Once that lot is depleted the process needs to be repeated to identify a fairly similar lot of serum.
Alternatively, you can rely on BPS Bioscience to perform the validation work on your behalf. At BPS Bioscience we have developed media tailored to our cell lines, with an optimized formulation for specific applications. In addition to animal serum validation, we have also determined the appropriate amount of antibiotics necessary for stable cell line maintenance (when relevant), thus providing a ready to use qualified tool for your cell culture. Trusting your cell culture to a suite of validated media will ensure a more reproducible and relevant outcome that will benefit your drug discovery process and, ultimately, benefit patients.
References
(1) Goet C., et al., 2022 Signal Transduction Immunohistochemistry: 197-212.
(2) Torres-Quesada O., et al., 2022 Cancers 14 (16): 3917.
(3) Wang J, et al., 2022 Biomolecules 12 (6): 743.
(4) Livnat G, et al., 2023 Int. J. Mol. Sci. 24(2): 1246.

