CRISPR-Based Synergistic Activation Mediator (SAM)
Introduction
Cas9 (Streptococcus pyogenes CRISPR-associated protein 9) is a sequence-specific prokaryotic endonuclease that is recruited to the DNA by a single guide RNA (sgRNA) and introduces a double stranded break into the DNA. This double stranded break is usually repaired through Non-Homologous End Joining (NHEJ) in mammalian cells (1). The target DNA sequence requires only 23 bp (base pairs) in length and should end with nucleotides GG. These are uncomplicated requirements, therefore sgRNAs can be designed to cleave any region of the genome. Mutant dCas9 (dead Cas9) has been engineered to disable the endonuclease activity, while still maintaining its ability to bind to guide RNA and unwind the targeted DNA strand without cleaving the DNA (2).
The CRISPR (SAM) system (CRISPR-based Synergistic Activation Mediator) is an astute combination of dCas9 and other molecular biology tools designed to activate the transcription of any endogenous gene of interest (reviewed in 3, 4, 5). The system comprises 3 components that form a DNA-binding complex once transfected into the cells. The first component is dCas9 fused to transcriptional activator VP64 (6, 7), typically composed of four tandem copies of VP16 (Herpes Simplex Viral Protein 16, amino acids 437-447) connected with glycine-serine linkers (8, 9). VP64 acts as a strong transcriptional activator when recruited by a DNA-binding protein.
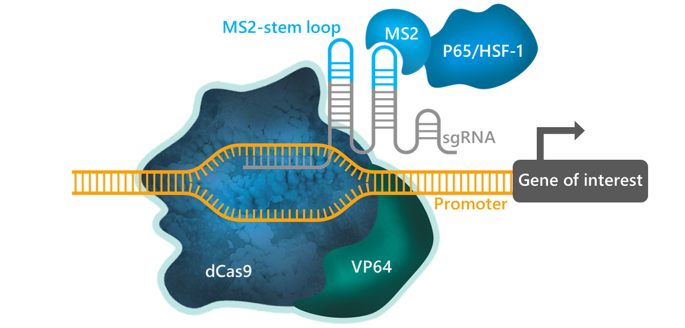
The other two components exploit the unique MS2 bacteriophage protein/RNA interaction system in which the coat protein of the bacteriophage binds tightly and specifically to a distinct 19-nucleotide RNA aptamer. In the second component of SAM, MS2 aptamers forming a characteristic stem loop structure are added to the sgRNA. The sgRNA-MS2 component forms a complex with dCas9 and directs it to the target DNA sequence next to the promoter region of the gene of interest. The sgRNA-MS2 aptamer recruits the third SAM component consisting of transcriptional activators P65 (Nuclear Factor NF-κB p65) and HSF1 (Heat Shock Factor 1) fused with an MS2-tag corresponding to the minimal aptamer-binding peptide of the MS2 coat protein (10). Once captured in the assembled complex at the gene promoter, P65 and HSF1 synergize with VP64 to robustly activate transcription of the downstream target gene by as much as a hundred-fold, depending on the gene. Theoretically, the SAM system can be used to target one or several gene promoters in the same cell.
CRISPR Activation (CRISPRa) Cell Lines
CRISPRa (SAM) cells stably express two of the required SAM components: the mutated dCas9 fused to VP64 and the p65/HSF1/MS2 tag construct. When the sgRNA-MS2 aptamer targeting the promoter of a gene of interest is introduced into the cells via electroporation, transfection, or transduction, dCas9-VP64 and MS2-P65-HSF1 are recruited to induce the transcription of the desired gene, resulting in high expression of the corresponding protein. These cell lines, including HEK293, HeLa, HepG2, Jurkat, MCF-7, and MDA-MB-231, represent a convenient and cost-effective platform to design activating experiments, needing only to introduce the desired sgRNA-MS2 construct.
MS2-sgRNA Plasmids and Lentiviruses
Replication-incompetent, integrating, VSV-G pseudotyped lentiviruses are ready to transduce SAM components in mammalian cells to generate new stable CRISPRa cells. The sgRNA-MS2 lentiviruses or sgRNA-MS2 plasmids contain a pool of 5 validated sgRNAs targeting the promoter region of a gene of interest, fused to MS2 aptamers and driven by a constitutive U6 promoter, and a puromycin resistance gene.
They are used in conjunction with constructs encoding dCas9-VP64 and MS2-P65-HSF1 (which contain blasticidin and hygromycin resistance genes, respectively) or with existing CRISPRa cell lines.
References:
(1) Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. (2012) 337: 816-821. PMID: 22745249.
(2) Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell (2013) 152: 1173-1183. PMID: 23452860.
(3) Adli M. The CRISPR tool kit for genome editing and beyond. Nat Commun. (2018) 9(1): 1911. PMID: 29765029
(4) Becirovic E. Maybe you can turn me on: CRISPRa-based strategies for therapeutic applications. Cell Mol Life Sci. 2022 Feb 12;79(2):130. doi: 10.1007/s00018-022-04175-8. PMID: 35152318.
(5) Nidhi S, et al. Novel CRISPR-Cas Systems: An Updated Review of the Current Achievements, Applications, and Future Research Perspectives. Int J Mol Sci. (2021) 22(7): 3327. PMID: 33805113.
(6) Maeder ML, et al. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. (2013) 10(10): 977-979. PMID: 23892898.
(7) Perez-Pinera P, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. (2013) 10(10): 973-976. PMID: 23892895.
(8) Seipel K, et al. Different activation domains stimulate transcription from remote ('enhancer') and proximal ('promoter') positions. EMBO J. (1992) 11(13): 4961-4968. PMID: 1464321.
(9) Beerli RR, et al. Toward controlling gene expression at will: Specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc Natl Acad Sci USA (1998); 95: 14628-14633. PMID: 9843940.
(10) Konermann S, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. (2015) 517: 583-588. PMID: 25494202.
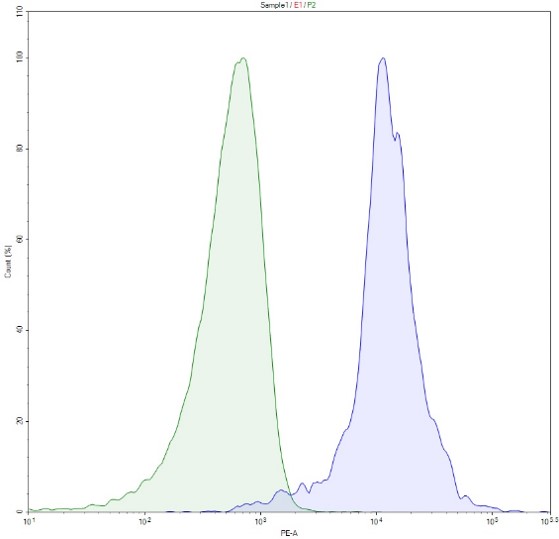
Induction of PD-1 in CRISPRa (SAM) Jurkat cells. Cells were electroporated with an sgRNA-MS2 plasmid targeting the PD-1 promoter (BPS Bioscience #78091). Cells were stained with PE-labeled anti-PD-1 antibody and analyzed by flow cytometry. Parental CRISPRa (SAM) Jurkat cells are shown in green, PD1-transfected cells are shown in blue.
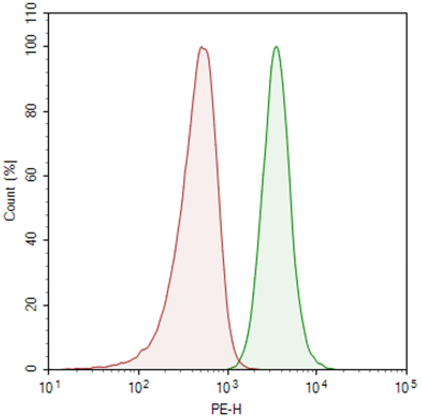
Induction of PD-1 in CRISPRa (SAM) MCF-7 cells. Cells were transduced with an sgRNA-MS2 Lentivirus targeting the human PD-1 promoter (BPS Bioscience #78190). Cells were stained with PE-labeled anti-PD-1 antibody (BioLegend #637309) and analyzed by flow cytometry. Parental CRISPRa (SAM) MCF7 cells are shown in red, and the PD-1 sgRNA-MS2-transduced cells are shown in green.

